Receptor tyrosine phosphatase-dependent cytoskeletal remodeling by the hedgehog-responsive gene MIM/BEG4
- PMID: 15684034
- PMCID: PMC2171717
- DOI: 10.1083/jcb.200409078
Receptor tyrosine phosphatase-dependent cytoskeletal remodeling by the hedgehog-responsive gene MIM/BEG4
Abstract
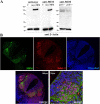
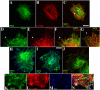
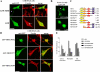
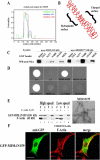
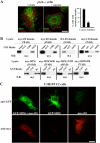
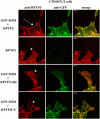
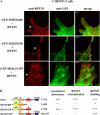
During development, dynamic remodeling of the actin cytoskeleton allows the precise placement and morphology of tissues. Morphogens such as Sonic hedgehog (Shh) and local cues such as receptor protein tyrosine phosphatases (RPTPs) mediate this process, but how they regulate the cytoskeleton is poorly understood. We previously identified Basal cell carcinoma-enriched gene 4 (BEG4)/Missing in Metastasis (MIM), a Shh-inducible, Wiskott-Aldrich homology 2 domain-containing protein that potentiates Gli transcription (Callahan, C.A., T. Ofstad, L. Horng, J.K. Wang, H.H. Zhen, P.A. Coulombe, and A.E. Oro. 2004. Genes Dev. 18:2724-2729). Here, we show that endogenous MIM is induced in a patched1-dependent manner and regulates the actin cytoskeleton. MIM functions by bundling F-actin, a process that requires self-association but is independent of G-actin binding. Cytoskeletal remodeling requires an activation domain distinct from sequences required for bundling in vitro. This domain associates with RPTPdelta and, in turn, enhances RPTPdelta membrane localization. MIM-dependent cytoskeletal changes can be inhibited using a soluble RPTPdelta-D2 domain. Our data suggest that the hedgehog-responsive gene MIM cooperates with RPTP to induce cytoskeletal changes.
Figures


Similar articles
-
MIM-B, a putative metastasis suppressor protein, binds to actin and to protein tyrosine phosphatase delta.Biochem J. 2003 Apr 15;371(Pt 2):463-71. doi: 10.1042/BJ20021962. Biochem J. 2003. PMID: 12570871 Free PMC article.
-
MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription.Genes Dev. 2004 Nov 15;18(22):2724-9. doi: 10.1101/gad.1221804. Genes Dev. 2004. PMID: 15545630 Free PMC article.
-
Murine missing in metastasis (MIM) mediates cell polarity and regulates the motility response to growth factors.PLoS One. 2011;6(6):e20845. doi: 10.1371/journal.pone.0020845. Epub 2011 Jun 9. PLoS One. 2011. PMID: 21695258 Free PMC article.
-
MIM: a multifunctional scaffold protein.J Mol Med (Berl). 2007 Jun;85(6):569-76. doi: 10.1007/s00109-007-0207-0. Epub 2007 May 12. J Mol Med (Berl). 2007. PMID: 17497115 Review.
-
LIM proteins: association with the actin cytoskeleton.Protoplasma. 2002 Feb;219(1-2):1-12. doi: 10.1007/s007090200000. Protoplasma. 2002. PMID: 11926060 Review.
Cited by
-
Tyrosine phosphatase PTPRD suppresses colon cancer cell migration in coordination with CD44.Exp Ther Med. 2011 May;2(3):457-463. doi: 10.3892/etm.2011.231. Epub 2011 Mar 21. Exp Ther Med. 2011. PMID: 22977525 Free PMC article.
-
Developmental expression and differentiation-related neuron-specific splicing of metastasis suppressor 1 (Mtss1) in normal and transformed cerebellar cells.BMC Dev Biol. 2007 Oct 9;7:111. doi: 10.1186/1471-213X-7-111. BMC Dev Biol. 2007. PMID: 17925019 Free PMC article.
-
Elevated MTSS1 expression associated with metastasis and poor prognosis of residual hepatitis B-related hepatocellular carcinoma.J Exp Clin Cancer Res. 2016 May 26;35(1):85. doi: 10.1186/s13046-016-0361-8. J Exp Clin Cancer Res. 2016. PMID: 27230279 Free PMC article.
-
MTSS1/Src family kinase dysregulation underlies multiple inherited ataxias.Proc Natl Acad Sci U S A. 2018 Dec 26;115(52):E12407-E12416. doi: 10.1073/pnas.1816177115. Epub 2018 Dec 7. Proc Natl Acad Sci U S A. 2018. PMID: 30530649 Free PMC article.
-
MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling.Dev Cell. 2010 Aug 17;19(2):270-83. doi: 10.1016/j.devcel.2010.07.009. Dev Cell. 2010. PMID: 20708589 Free PMC article.
References
-
- Bear, J.E., M. Krause, and F.B. Gertler. 2001. Regulating cellular actin assembly. Curr. Opin. Cell Biol. 13:158–166. - PubMed
-
- Bretscher, A., and K. Weber. 1980. Villin is a major protein of the microvillus cytoskeleton which binds both G and F actin in a calcium-dependent manner. Cell. 20:839–847. - PubMed
-
- Charron, F., E. Stein, J. Jeong, A.P. McMahon, and M. Tessier-Lavigne. 2003. The morphogen sonic hedgehog is an axonal chemoattractant that collaborates with netrin-1 in midline axon guidance. Cell. 113:11–23. - PubMed
-
- Dent, E.W., F. Tang, and K. Kalil. 2003. Axon guidance by growth cones and branches: common cytoskeletal and signaling mechanisms. Neuroscientist. 9:343–353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

