Inhibition of coxsackievirus B3 replication by small interfering RNAs requires perfect sequence match in the central region of the viral positive strand
- PMID: 15681418
- PMCID: PMC546545
- DOI: 10.1128/JVI.79.4.2151-2159.2005
Inhibition of coxsackievirus B3 replication by small interfering RNAs requires perfect sequence match in the central region of the viral positive strand
Abstract
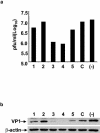
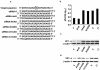
Coxsackievirus B3 (CVB3) is the most common causal agent of viral myocarditis, but existing drug therapies are of limited value. Application of small interfering RNA (siRNA) in knockdown of gene expression is an emerging technology in antiviral gene therapy. To investigate whether RNA interference (RNAi) can protect against CVB3 infection, we evaluated the effects of RNAi on viral replication in HeLa cells and murine cardiomyocytes by using five CVB3-specific siRNAs targeting distinct regions of the viral genome. The most effective one is siRNA-4, targeting the viral protease 2A, achieving a 92% inhibition of CVB3 replication. The specific RNAi effects could last at least 48 h, and cell viability assay revealed that 90% of siRNA-4-pretreated cells were still alive and lacked detectable viral protein expression 48 h postinfection. Moreover, administration of siRNAs after viral infection could also effectively inhibit viral replication, indicating its therapeutic potential. Further evaluation by combination found that no enhanced inhibitory effects were observed when siRNA-4 was cotransfected with each of the other four candidates. In mutational analysis of the mechanisms of siRNA action, we found that siRNA functions by targeting the positive strand of virus and requires a perfect sequence match in the central region of the target, but mismatches were more tolerated near the 3' end than the 5' end of the antisense strand. These findings reveal an effective target for CVB3 silencing and provide a new possibility for antiviral intervention.
Figures


Similar articles
-
Expression of short hairpin RNAs against the coxsackievirus B3 exerts potential antiviral effects in Cos-7 cells and in mice.Virus Res. 2007 Apr;125(1):9-13. doi: 10.1016/j.virusres.2006.11.009. Epub 2007 Jan 12. Virus Res. 2007. PMID: 17222937
-
Developing an effective RNA interference strategy against a plus-strand RNA virus: silencing of coxsackievirus B3 and its cognate coxsackievirus-adenovirus receptor.Biol Chem. 2005 Sep;386(9):857-63. doi: 10.1515/BC.2005.100. Biol Chem. 2005. PMID: 16164410
-
[Short interfering RNA-mediated inhibition of coxsakievirus B3 infection in vitro].Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2007 Jun;21(2):150-2. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2007. PMID: 17653320 Chinese.
-
Control of HIV-1 replication by RNA interference.Virus Res. 2004 Jun 1;102(1):53-8. doi: 10.1016/j.virusres.2004.01.015. Virus Res. 2004. PMID: 15068880 Review.
-
A Kidnapping Story: How Coxsackievirus B3 and Its Host Cell Interact.Cell Physiol Biochem. 2019;53(1):121-140. doi: 10.33594/000000125. Cell Physiol Biochem. 2019. PMID: 31230428 Review.
Cited by
-
siRNA efficiency: structure or sequence-that is the question.J Biomed Biotechnol. 2006;2006(4):83757. doi: 10.1155/JBB/2006/83757. J Biomed Biotechnol. 2006. PMID: 17057371 Free PMC article.
-
Exploiting the therapeutic potential of microRNAs in viral diseases: expectations and limitations.Mol Diagn Ther. 2010 Oct 1;14(5):271-82. doi: 10.1007/BF03256383. Mol Diagn Ther. 2010. PMID: 21053993 Free PMC article. Review.
-
Broad-spectrum antiviral activity of small interfering RNA targeting the conserved RNA termini of Lassa virus.Antimicrob Agents Chemother. 2007 Jun;51(6):2215-8. doi: 10.1128/AAC.01368-06. Epub 2007 Mar 19. Antimicrob Agents Chemother. 2007. PMID: 17371814 Free PMC article.
-
Coxsackievirus B3 and adenovirus infections of cardiac cells are efficiently inhibited by vector-mediated RNA interference targeting their common receptor.Gene Ther. 2007 Jun;14(12):960-71. doi: 10.1038/sj.gt.3302948. Epub 2007 Mar 22. Gene Ther. 2007. PMID: 17377597 Free PMC article.
-
Therapeutic opportunities of small interfering RNA.Fundam Clin Pharmacol. 2009 Aug;23(4):367-86. doi: 10.1111/j.1472-8206.2009.00694.x. Fundam Clin Pharmacol. 2009. PMID: 19709318 Free PMC article. Review.
References
-
- Baulcombe, D. 2002. RNA silencing. Curr. Biol. 12:R82—R84. - PubMed
-
- Bertrand, J. R., M. Pottier, A. Vekris, P. Opolon, A. Maksimenko, and C. Malvy. 2002. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem. Biophys. Res. Commun. 296:1000-1004. - PubMed
-
- Cetta, F., and V. V. Michels. 1995. The autoimmune basis of dilated cardiomyopathy. Ann. Med. 27:169-173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

