Genetic structure and diversity in Oryza sativa L
- PMID: 15654106
- PMCID: PMC1449546
- DOI: 10.1534/genetics.104.035642
Genetic structure and diversity in Oryza sativa L
Abstract
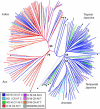
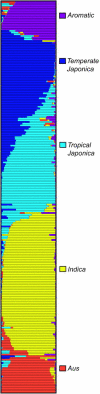
The population structure of domesticated species is influenced by the natural history of the populations of predomesticated ancestors, as well as by the breeding system and complexity of the breeding practices exercised by humans. Within Oryza sativa, there is an ancient and well-established divergence between the two major subspecies, indica and japonica, but finer levels of genetic structure are suggested by the breeding history. In this study, a sample of 234 accessions of rice was genotyped at 169 nuclear SSRs and two chloroplast loci. The data were analyzed to resolve the genetic structure and to interpret the evolutionary relationships between groups. Five distinct groups were detected, corresponding to indica, aus, aromatic, temperate japonica, and tropical japonica rices. Nuclear and chloroplast data support a closer evolutionary relationship between the indica and the aus and among the tropical japonica, temperate japonica, and aromatic groups. Group differences can be explained through contrasting demographic histories. With the availability of rice genome sequence, coupled with a large collection of publicly available genetic resources, it is of interest to develop a population-based framework for the molecular analysis of diversity in O. sativa.
Figures
Similar articles
-
Chloroplast phylogeography of AA genome rice species.Mol Phylogenet Evol. 2018 Oct;127:475-487. doi: 10.1016/j.ympev.2018.05.002. Epub 2018 May 16. Mol Phylogenet Evol. 2018. PMID: 29753711
-
Genetic diversity and structure in hill rice (Oryza sativa L.) landraces from the North-Eastern Himalayas of India.BMC Genet. 2016 Jul 13;17(1):107. doi: 10.1186/s12863-016-0414-1. BMC Genet. 2016. PMID: 27412613 Free PMC article.
-
Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa.Proc Natl Acad Sci U S A. 2006 Jun 20;103(25):9578-83. doi: 10.1073/pnas.0603152103. Epub 2006 Jun 9. Proc Natl Acad Sci U S A. 2006. PMID: 16766658 Free PMC article.
-
The complex history of the domestication of rice.Ann Bot. 2007 Nov;100(5):951-7. doi: 10.1093/aob/mcm128. Epub 2007 Jul 6. Ann Bot. 2007. PMID: 17617555 Free PMC article. Review.
-
Genome-wide intraspecific DNA-sequence variations in rice.Curr Opin Plant Biol. 2003 Apr;6(2):134-8. doi: 10.1016/s1369-5266(03)00004-9. Curr Opin Plant Biol. 2003. PMID: 12667869 Review.
Cited by
-
Copy number variation of the restorer Rf4 underlies human selection of three-line hybrid rice breeding.Nat Commun. 2023 Nov 13;14(1):7333. doi: 10.1038/s41467-023-43009-4. Nat Commun. 2023. PMID: 37957162 Free PMC article.
-
Oryza CLIMtools: A genome-environment association resource reveals adaptive roles for heterotrimeric G proteins in the regulation of rice agronomic traits.Plant Commun. 2024 Apr 8;5(4):100813. doi: 10.1016/j.xplc.2024.100813. Epub 2024 Jan 11. Plant Commun. 2024. PMID: 38213027 Free PMC article.
-
Whole-genome analysis revealed the positively selected genes during the differentiation of indica and temperate japonica rice.PLoS One. 2015 Mar 16;10(3):e0119239. doi: 10.1371/journal.pone.0119239. eCollection 2015. PLoS One. 2015. PMID: 25774680 Free PMC article.
-
Homologues of CsLOB1 in citrus function as disease susceptibility genes in citrus canker.Mol Plant Pathol. 2017 Aug;18(6):798-810. doi: 10.1111/mpp.12441. Epub 2016 Aug 11. Mol Plant Pathol. 2017. PMID: 27276658 Free PMC article.
-
Genetic Diversity and Population Structure in Aromatic and Quality Rice (Oryza sativa L.) Landraces from North-Eastern India.PLoS One. 2015 Jun 12;10(6):e0129607. doi: 10.1371/journal.pone.0129607. eCollection 2015. PLoS One. 2015. PMID: 26067999 Free PMC article.
References
-
- Ahuja, S. C., D. V. S. Panwar, U. Ahuja and K. R. Gupta, 1995 Basmati Rice: The Scented Pearl. Directorate of Publications, C.C.S. Haryana Agricultural University, Hisar, Haryana, India.
-
- Cai, H. W., and H. Morishima, 2002. QTL clusters reflect character associations in wild and cultivated rice. Theor. Appl. Genet. 104: 1217–1228. - PubMed
-
- Chen, X., S. Temnykh, Y. Xu, Y. G. Cho and S. R. McCouch, 1997. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor. Appl. Genet. 95: 553–567.
-
- Cheng, C. Y., R. Motohashi, S. Tsuchimoto, Y. Fukuta, H. Ohtsubo et al., 2003. Polyphyletic origin of cultivated rice: based on the interspersion pattern of SINEs. Mol. Biol. Evol. 20: 67–75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

