Subtle mutational changes in the SU protein of a natural feline leukemia virus subgroup A isolate alter disease spectrum
- PMID: 15650161
- PMCID: PMC544135
- DOI: 10.1128/JVI.79.3.1351-1360.2005
Subtle mutational changes in the SU protein of a natural feline leukemia virus subgroup A isolate alter disease spectrum
Abstract
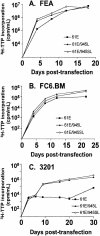
FeLV-945 is a representative isolate of the natural feline leukemia virus (FeLV) variant predominant in non-T-cell malignant, proliferative, and degenerative diseases in a geographic cohort. The FeLV-945 surface glycoprotein (SU) is closely related to natural horizontally transmissible FeLV subgroup A (FeLV-A) but was found to differ from a prototype to a larger extent than the members of FeLV-A differ among themselves. The sequence differences included point mutations restricted largely to the functional domains of SU, i.e., VRA, VRB, and PRR. Despite the sequence differences in these critical domains, measurements of receptor utilization, including host range and superinfection interference, confirmed the assignment of FeLV-945 to subgroup A. Other proviruses isolated from the cohort contained similar sequence hallmarks and were assigned to FeLV subgroup A. A provirus from cat 1046 contained a histidine-to-proline change at SU residue 6 within an SPHQ motif that was previously identified as a critical mediator of fusion events during virus entry. The 1046 pseudotype virus entered cells only in the presence of the soluble cofactor FeLIX provided in trans, but it retained an ecotropic host range even in the presence of FeLIX. The mutational changes in FeLV-945 were shown to confer significant functional differences compared to prototype FeLV-A viruses. The substitution of FeLV-945 envelope gene sequences for FeLV-A/61E sequences conferred a small but statistically significant replicative advantage in some feline cells. Moreover, substitution of the unique FeLV-945 long terminal repeat and envelope gene for those of FeLV-A/61E altered the disease spectrum entirely, from a thymic lymphoma of a T-cell origin to an as yet uncharacterized multicentric lymphoma that did not contain T cells.
Figures



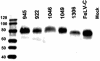
Similar articles
-
Unique long terminal repeat and surface glycoprotein gene sequences of feline leukemia virus as determinants of disease outcome.J Virol. 2005 May;79(9):5278-87. doi: 10.1128/JVI.79.9.5278-5287.2005. J Virol. 2005. PMID: 15827142 Free PMC article.
-
The surface glycoprotein of feline leukemia virus isolate FeLV-945 is a determinant of altered pathogenesis in the presence or absence of the unique viral long terminal repeat.J Virol. 2013 Oct;87(19):10874-83. doi: 10.1128/JVI.01130-13. Epub 2013 Jul 31. J Virol. 2013. PMID: 23903838 Free PMC article.
-
Advances in understanding molecular determinants in FeLV pathology.Vet Immunol Immunopathol. 2008 May 15;123(1-2):14-22. doi: 10.1016/j.vetimm.2008.01.008. Epub 2008 Jan 19. Vet Immunol Immunopathol. 2008. PMID: 18289704 Free PMC article. Review.
-
The surface glycoprotein of a natural feline leukemia virus subgroup A variant, FeLV-945, as a determinant of disease outcome.Vet Immunol Immunopathol. 2011 Oct 15;143(3-4):221-6. doi: 10.1016/j.vetimm.2011.06.015. Epub 2011 Jun 12. Vet Immunol Immunopathol. 2011. PMID: 21764142 Free PMC article.
-
Cooperating events in lymphomagenesis mediated by feline leukemia virus.Leukemia. 1997 Apr;11 Suppl 3:239-41. Leukemia. 1997. PMID: 9209353 Review.
Cited by
-
Identification of novel subgroup A variants with enhanced receptor binding and replicative capacity in primary isolates of anaemogenic strains of feline leukaemia virus.Retrovirology. 2012 May 31;9:48. doi: 10.1186/1742-4690-9-48. Retrovirology. 2012. PMID: 22650160 Free PMC article.
-
Unique long terminal repeat and surface glycoprotein gene sequences of feline leukemia virus as determinants of disease outcome.J Virol. 2005 May;79(9):5278-87. doi: 10.1128/JVI.79.9.5278-5287.2005. J Virol. 2005. PMID: 15827142 Free PMC article.
-
A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A.J Virol. 2006 Apr;80(7):3378-85. doi: 10.1128/JVI.80.7.3378-3385.2006. J Virol. 2006. PMID: 16537605 Free PMC article.
-
Feline leukemia virus and other pathogens as important threats to the survival of the critically endangered Iberian lynx (Lynx pardinus).PLoS One. 2009;4(3):e4744. doi: 10.1371/journal.pone.0004744. Epub 2009 Mar 9. PLoS One. 2009. PMID: 19270739 Free PMC article.
-
Viral determinants of FeLV infection and pathogenesis: lessons learned from analysis of a natural cohort.Viruses. 2011 Sep;3(9):1681-98. doi: 10.3390/v3091681. Epub 2011 Sep 9. Viruses. 2011. PMID: 21994802 Free PMC article. Review.
References
-
- Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. - PubMed
-
- Athas, G. B., B. Choi, S. Prabhu, P. A. Lobelle-Rich, and L. S. Levy. 1995. Genetic determinants of feline leukemia virus-induced multicentric lymphomas. Virology 214:431-438. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

