CD63 interacts with the carboxy terminus of the colonic H+-K+-ATPase to decrease [corrected] plasma membrane localization and 86Rb+ uptake
- PMID: 15647390
- PMCID: PMC1868892
- DOI: 10.1152/ajpcell.00463.2004
CD63 interacts with the carboxy terminus of the colonic H+-K+-ATPase to decrease [corrected] plasma membrane localization and 86Rb+ uptake
Erratum in
- Am J Physiol Cell Physiol. 2007 Apr;292(4):C1567
Abstract
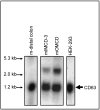
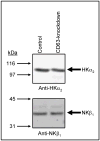
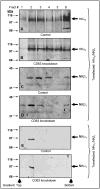
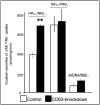
The carboxy terminus (CT) of the colonic H(+)-K(+)-ATPase is required for stable assembly with the beta-subunit, translocation to the plasma membrane, and efficient function of the transporter. To identify protein-protein interactions involved in the localization and function of HKalpha(2), we selected 84 amino acids in the CT of the alpha-subunit of mouse colonic H(+)-K(+)-ATPase (CT-HKalpha(2)) as the bait in a yeast two-hybrid screen of a mouse kidney cDNA library. The longest identified clone was CD63. To characterize the interaction of CT-HKalpha(2) with CD63, recombinant CT-HKalpha(2) and CD63 were synthesized in vitro and incubated, and complexes were immunoprecipitated. CT-HKalpha(2) protein (but not CT-HKalpha(1)) coprecipitated with CD63, confirming stable assembly of HKalpha(2) with CD63. In HEK-293 transfected with HKalpha(2) plus beta(1)-Na(+)-K(+)-ATPase, suppression of CD63 by RNA interference increased cell surface expression of HKalpha(2)/NKbeta(1) and (86)Rb(+) uptake. These studies demonstrate that CD63 participates in the regulation of the abundance of the HKalpha(2)-NKbeta(1) complex in the cell membrane.
Figures

Similar articles
-
The carboxy terminus of the colonic H(+), K(+)-ATPase alpha-subunit is required for stable beta subunit assembly and function.Kidney Int. 2004 Apr;65(4):1301-10. doi: 10.1111/j.1523-1755.2004.00507.x. Kidney Int. 2004. PMID: 15086469
-
The effect of beta-subunit assembly on function and localization of the colonic H+,K+-ATPase alpha-subunit.Kidney Int. 2004 Sep;66(3):1068-75. doi: 10.1111/j.1523-1755.2004.00856.x. Kidney Int. 2004. PMID: 15327400
-
Molecular regulation and physiology of the H+,K+ -ATPases in kidney.Semin Nephrol. 2006 Sep;26(5):345-51. doi: 10.1016/j.semnephrol.2006.07.003. Semin Nephrol. 2006. PMID: 17071328 Review.
-
Palytoxin acts on Na(+),K (+)-ATPase but not nongastric H(+),K (+)-ATPase.J Membr Biol. 2007 Apr;216(2-3):107-16. doi: 10.1007/s00232-007-9040-1. Epub 2007 Jul 17. J Membr Biol. 2007. PMID: 17639367 Free PMC article.
-
The renal H+-K+-ATPases: physiology, regulation, and structure.Am J Physiol Renal Physiol. 2010 Jan;298(1):F12-21. doi: 10.1152/ajprenal.90723.2008. Epub 2009 Jul 29. Am J Physiol Renal Physiol. 2010. PMID: 19640897 Free PMC article. Review.
Cited by
-
Deficiency of the tetraspanin CD63 associated with kidney pathology but normal lysosomal function.Mol Cell Biol. 2009 Feb;29(4):1083-94. doi: 10.1128/MCB.01163-08. Epub 2008 Dec 15. Mol Cell Biol. 2009. PMID: 19075008 Free PMC article.
-
CD63 is not required for production of infectious human immunodeficiency virus type 1 in human macrophages.J Virol. 2008 May;82(10):4751-61. doi: 10.1128/JVI.02320-07. Epub 2008 Mar 5. J Virol. 2008. PMID: 18321974 Free PMC article.
-
pH-dependent regulation of the α-subunit of H+-K+-ATPase (HKα2).Am J Physiol Renal Physiol. 2011 Sep;301(3):F536-43. doi: 10.1152/ajprenal.00220.2011. Epub 2011 Jun 8. Am J Physiol Renal Physiol. 2011. PMID: 21653633 Free PMC article.
-
Palmitoylation-dependent association with CD63 targets the Ca2+ sensor synaptotagmin VII to lysosomes.J Cell Biol. 2010 Nov 1;191(3):599-613. doi: 10.1083/jcb.201003021. J Cell Biol. 2010. PMID: 21041449 Free PMC article.
References
-
- Abramowitz J, Mattera R, Liao CF, Olate J, Perez-Ripoll E, Birnbaumer L, Codina J. Screening of cDNA libraries with oligonucleotides as applied to signal transducing G proteins, receptors and effectors. J Recept Res. 1988;8:561–588. - PubMed
-
- Alper SL. Genetic diseases of acid-base transporters. Annu Rev Physiol. 2002;64:899–923. - PubMed
-
- Arystarkhova E, Sweadner KJ. Tissue-specific expression of the Na+,K+-ATPase β3-subunit. The presence of β3 in lung and liver addresses the problem of the missing subunit. J Biol Chem. 1997;272:22405–22408. - PubMed
-
- Asano S, Hoshina S, Nakaie Y, Watanabe T, Sato M, Suzuki Y, Takeguchi N. Functional expression of putative H+,K+-ATPase from guinea pig distal colon. Am J Physiol. 1998;275:C669–674. - PubMed
-
- Berditchevski F, Gilbert E, Griffiths MR, Fitter S, Ashman L, Jenner SJ. Analysis of the CD151-α3β1 integrin and CD151-tetraspanin interactions by mutagenesis. J Biol Chem. 2001;276:41165–41174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

