Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage
- PMID: 15601841
- PMCID: PMC538766
- DOI: 10.1128/MCB.25.1.185-196.2005
Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage
Abstract
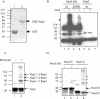
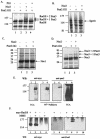
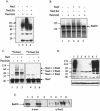
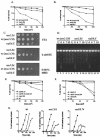
The Schizosaccharomyces pombe SMC proteins Rad18 (Smc6) and Spr18 (Smc5) exist in a high-M(r) complex which also contains the non-SMC proteins Nse1, Nse2, Nse3, and Rad62. The Smc5-6 complex, which is essential for viability, is required for several aspects of DNA metabolism, including recombinational repair and maintenance of the DNA damage checkpoint. We have characterized Nse2 and show here that it is a SUMO ligase. Smc6 (Rad18) and Nse3, but not Smc5 (Spr18) or Nse1, are sumoylated in vitro in an Nse2-dependent manner, and Nse2 is itself autosumoylated, predominantly on the C-terminal part of the protein. Mutations of C195 and H197 in the Nse2 RING-finger-like motif abolish Nse2-dependent sumoylation. nse2.SA mutant cells, in which nse2.C195S-H197A is integrated as the sole copy of nse2, are viable, whereas the deletion of nse2 is lethal. Smc6 (Rad18) is sumoylated in vivo: the sumoylation level is increased upon exposure to DNA damage and is drastically reduced in the nse2.SA strain. Since nse2.SA cells are sensitive to DNA-damaging agents and to exposure to hydroxyurea, this implicates the Nse2-dependent sumoylation activity in DNA damage responses but not in the essential function of the Smc5-6 complex.
Figures


Similar articles
-
Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex.Mol Cell Biol. 2005 Jan;25(1):172-84. doi: 10.1128/MCB.25.1.172-184.2005. Mol Cell Biol. 2005. PMID: 15601840 Free PMC article.
-
Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis.Mol Biol Cell. 2004 Nov;15(11):4866-76. doi: 10.1091/mbc.e04-05-0436. Epub 2004 Aug 25. Mol Biol Cell. 2004. PMID: 15331764 Free PMC article.
-
Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex.J Biol Chem. 2003 Nov 14;278(46):45460-7. doi: 10.1074/jbc.M308828200. Epub 2003 Sep 8. J Biol Chem. 2003. PMID: 12966087
-
A new SUMO ligase in the DNA damage response.DNA Repair (Amst). 2006 Jan 5;5(1):138-41. doi: 10.1016/j.dnarep.2005.08.003. Epub 2005 Sep 27. DNA Repair (Amst). 2006. PMID: 16198156 Review.
-
The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability.FEBS Lett. 2011 Sep 16;585(18):2907-13. doi: 10.1016/j.febslet.2011.04.067. Epub 2011 May 4. FEBS Lett. 2011. PMID: 21550342 Review.
Cited by
-
SUMO chain formation is required for response to replication arrest in S. pombe.PLoS One. 2009 Aug 25;4(8):e6750. doi: 10.1371/journal.pone.0006750. PLoS One. 2009. PMID: 19707600 Free PMC article.
-
NMR metabolomic profiling reveals new roles of SUMOylation in DNA damage response.J Proteome Res. 2010 Oct 1;9(10):5382-8. doi: 10.1021/pr100614a. J Proteome Res. 2010. PMID: 20695451 Free PMC article.
-
Depletion of SMC5/6 sensitizes male germ cells to DNA damage.Mol Biol Cell. 2018 Dec 1;29(25):3003-3016. doi: 10.1091/mbc.E18-07-0459. Epub 2018 Oct 3. Mol Biol Cell. 2018. PMID: 30281394 Free PMC article.
-
Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex.Mol Cell. 2009 Sep 11;35(5):657-68. doi: 10.1016/j.molcel.2009.06.032. Mol Cell. 2009. PMID: 19748359 Free PMC article.
-
The S phase checkpoint promotes the Smc5/6 complex dependent SUMOylation of Pol2, the catalytic subunit of DNA polymerase ε.PLoS Genet. 2019 Nov 25;15(11):e1008427. doi: 10.1371/journal.pgen.1008427. eCollection 2019 Nov. PLoS Genet. 2019. PMID: 31765407 Free PMC article.
References
-
- al-Khodairy, F., T. Enoch, I. M. Hagan, and A. M. Carr. 1995. The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J. Cell Sci. 108:475-486. - PubMed
-
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner, and S. J. Elledge. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9:1169-1182. - PubMed
-
- Barbet, N. C., and A. M. Carr. 1993. Fission yeast wee1 protein kinase is not required for DNA damage-dependent mitotic arrest. Nature 364:824-827. - PubMed
-
- Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. D. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345-356. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
