The carboxyl-terminal region of human cytomegalovirus IE1491aa contains an acidic domain that plays a regulatory role and a chromatin-tethering domain that is dispensable during viral replication
- PMID: 15596818
- PMCID: PMC538725
- DOI: 10.1128/JVI.79.1.225-233.2005
The carboxyl-terminal region of human cytomegalovirus IE1491aa contains an acidic domain that plays a regulatory role and a chromatin-tethering domain that is dispensable during viral replication
Abstract
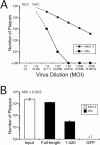
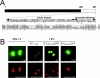
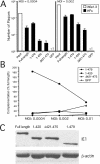
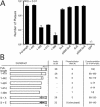
The human cytomegalovirus major immediate-early (alpha) protein IE1(491aa) plays an important role in controlling viral gene expression at low multiplicities of infection. With a transient complementation assay, full-length IE1(491aa) enhanced the growth of ie1 mutant virus CR208 20-fold better than a deletion mutant lacking 71 carboxyl-terminal amino acids (IE1(1-420aa)). A 16-amino-acid domain between amino acids 476 and 491 was both necessary and sufficient for chromatin-tethering activity; however, this domain was completely dispensable for complementation of CR208 replication. The proximal 55-amino-acid acidic domain (amino acids 421 to 475) was found to be most important for function. A deletion mutant lacking only this domain retained chromatin-tethering activity but failed to complement mutant virus. Interestingly, serine phosphorylation (at amino acids 399, 402, 406, 423, 428, 431, 448, 451, and 455) was not required for complementation. These results show that IE1(491aa) is composed of at least two domains that support replication, a region located between amino acids 1 and 399 that complements ie1 mutant virus replication to low levels and an acidic domain between amino acids 421 and 479 that dramatically enhances complementation.
Figures
Similar articles
-
A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation.Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11321-6. doi: 10.1073/pnas.93.21.11321. Proc Natl Acad Sci U S A. 1996. PMID: 8876134 Free PMC article.
-
Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant.J Virol. 1998 Jan;72(1):366-79. doi: 10.1128/JVI.72.1.366-379.1998. J Virol. 1998. PMID: 9420235 Free PMC article.
-
The chromatin-tethering domain of human cytomegalovirus immediate-early (IE) 1 mediates associations of IE1, PML and STAT2 with mitotic chromosomes, but is not essential for viral replication.J Gen Virol. 2012 Apr;93(Pt 4):716-721. doi: 10.1099/vir.0.037986-0. Epub 2011 Dec 7. J Gen Virol. 2012. PMID: 22158879
-
SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication.J Virol. 2004 Jul;78(14):7803-12. doi: 10.1128/JVI.78.14.7803-7812.2004. J Virol. 2004. PMID: 15220454 Free PMC article.
-
Exon 3 of the human cytomegalovirus major immediate-early region is required for efficient viral gene expression and for cellular cyclin modulation.J Virol. 2005 Jun;79(12):7438-52. doi: 10.1128/JVI.79.12.7438-7452.2005. J Virol. 2005. PMID: 15919900 Free PMC article.
Cited by
-
Revisiting promyelocytic leukemia protein targeting by human cytomegalovirus immediate-early protein 1.PLoS Pathog. 2020 May 4;16(5):e1008537. doi: 10.1371/journal.ppat.1008537. eCollection 2020 May. PLoS Pathog. 2020. PMID: 32365141 Free PMC article.
-
Human Cytomegalovirus Immediate-Early 1 Protein Rewires Upstream STAT3 to Downstream STAT1 Signaling Switching an IL6-Type to an IFNγ-Like Response.PLoS Pathog. 2016 Jul 7;12(7):e1005748. doi: 10.1371/journal.ppat.1005748. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27387064 Free PMC article.
-
Contribution of GADD45 family members to cell death suppression by cellular Bcl-xL and cytomegalovirus vMIA.J Virol. 2005 Dec;79(23):14923-32. doi: 10.1128/JVI.79.23.14923-14932.2005. J Virol. 2005. PMID: 16282491 Free PMC article.
-
The carboxy terminal region of the human cytomegalovirus immediate early 1 (IE1) protein disrupts type II inteferon signaling.Viruses. 2014 Apr 2;6(4):1502-24. doi: 10.3390/v6041502. Viruses. 2014. PMID: 24699362 Free PMC article.
-
Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis.J Virol. 2005 Oct;79(19):12205-17. doi: 10.1128/JVI.79.19.12205-12217.2005. J Virol. 2005. PMID: 16160147 Free PMC article.
References
-
- Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. - PubMed
-
- Berk, A. J., T. G. Boyer, A. N. Kapanidis, R. H. Ebright, N. N. Kobayashi, P. J. Horn, S. M. Sullivan, R. Koop, M. A. Surby, and S. J. Triezenberg. 1998. Mechanisms of viral activators. Cold Spring Harb. Symp. Quant. Biol. 63:243-252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

