Substitution of feline leukemia virus long terminal repeat sequences into murine leukemia virus alters the pattern of insertional activation and identifies new common insertion sites
- PMID: 15596801
- PMCID: PMC538733
- DOI: 10.1128/JVI.79.1.57-66.2005
Substitution of feline leukemia virus long terminal repeat sequences into murine leukemia virus alters the pattern of insertional activation and identifies new common insertion sites
Abstract
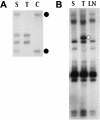
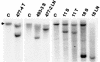
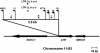
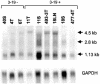
The recombinant retrovirus, MoFe2-MuLV (MoFe2), was constructed by replacing the U3 region of Moloney murine leukemia virus (M-MuLV) with homologous sequences from the FeLV-945 LTR. NIH/Swiss mice neonatally inoculated with MoFe2 developed T-cell lymphomas of immature thymocyte surface phenotype. MoFe2 integrated infrequently (0 to 9%) near common insertion sites (CISs) previously identified for either parent virus. Using three different strategies, CISs in MoFe2-induced tumors were identified at six loci, none of which had been previously reported as CISs in tumors induced by either parent virus in wild-type animals. Two of the newly identified CISs had not previously been implicated in lymphoma in any retrovirus model. One of these, designated 3-19, encodes the p101 regulatory subunit of phosphoinositide-3-kinase-gamma. The other, designated Rw1, is predicted to encode a protein that functions in the immune response to virus infection. Thus, substitution of FeLV-945 U3 sequences into the M-MuLV long terminal repeat (LTR) did not alter the target tissue for M-MuLV transformation but significantly altered the pattern of CIS utilization in the induction of T-cell lymphoma. These observations support a growing body of evidence that the distinctive sequence and/or structure of the retroviral LTR determines its pattern of insertional activation. The findings also demonstrate the oligoclonal nature of retrovirus-induced lymphomas by demonstrating proviral insertions at CISs in subdominant populations in the tumor mass. Finally, the findings demonstrate the utility of novel recombinant retroviruses such as MoFe2 to contribute new genes potentially relevant to the induction of lymphoid malignancy.
Figures
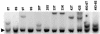


Similar articles
-
Tumorigenic potential of a recombinant retrovirus containing sequences from Moloney murine leukemia virus and feline leukemia virus.J Virol. 1998 Feb;72(2):1078-84. doi: 10.1128/JVI.72.2.1078-1084.1998. J Virol. 1998. PMID: 9445002 Free PMC article.
-
Analysis of the disease potential of a recombinant retrovirus containing Friend murine leukemia virus sequences and a unique long terminal repeat from feline leukemia virus.J Virol. 2002 Feb;76(3):1527-32. doi: 10.1128/jvi.76.3.1527-1532.2002. J Virol. 2002. PMID: 11773427 Free PMC article.
-
Genetic determinants of feline leukemia virus-induced multicentric lymphomas.Virology. 1995 Dec 20;214(2):431-8. doi: 10.1006/viro.1995.0053. Virology. 1995. PMID: 8553544
-
Advances in understanding molecular determinants in FeLV pathology.Vet Immunol Immunopathol. 2008 May 15;123(1-2):14-22. doi: 10.1016/j.vetimm.2008.01.008. Epub 2008 Jan 19. Vet Immunol Immunopathol. 2008. PMID: 18289704 Free PMC article. Review.
-
Molecular pathogenesis of feline leukemia virus-induced malignancies: insertional mutagenesis.Vet Immunol Immunopathol. 2008 May 15;123(1-2):138-43. doi: 10.1016/j.vetimm.2008.01.019. Epub 2008 Jan 19. Vet Immunol Immunopathol. 2008. PMID: 18313764 Review.
Cited by
-
Radiation leukemia virus common integration at the Kis2 locus: simultaneous overexpression of a novel noncoding RNA and of the proximal Phf6 gene.J Virol. 2005 Sep;79(17):11443-56. doi: 10.1128/JVI.79.17.11443-11456.2005. J Virol. 2005. PMID: 16103195 Free PMC article.
-
Silencing of IRF8 Mediated by m6A Modification Promotes the Progression of T-Cell Acute Lymphoblastic Leukemia.Adv Sci (Weinh). 2023 Jan;10(2):e2201724. doi: 10.1002/advs.202201724. Epub 2022 Dec 7. Adv Sci (Weinh). 2023. PMID: 36478193 Free PMC article.
-
Unique long terminal repeat and surface glycoprotein gene sequences of feline leukemia virus as determinants of disease outcome.J Virol. 2005 May;79(9):5278-87. doi: 10.1128/JVI.79.9.5278-5287.2005. J Virol. 2005. PMID: 15827142 Free PMC article.
-
Diminished potential for B-lymphoid differentiation after murine leukemia virus infection in vivo and in EML hematopoietic progenitor cells.J Virol. 2007 Jul;81(13):7274-9. doi: 10.1128/JVI.00250-07. Epub 2007 Apr 11. J Virol. 2007. PMID: 17428873 Free PMC article.
-
Feline leukemia virus integrase and capsid packaging functions do not change the insertion profile of standard Moloney retroviral vectors.Gene Ther. 2010 Jun;17(6):799-804. doi: 10.1038/gt.2010.24. Epub 2010 Mar 18. Gene Ther. 2010. PMID: 20237508 Free PMC article.
References
-
- Athas, G. B., B. Choi, S. Prabhu, P. A. Lobelle-Rich, and L. S. Levy. 1995. Genetic determinants of feline leukemia virus-induced multicentric lymphomas. Virology 214:431-438. - PubMed
-
- Baier, R., T. Bondeva, R. Klinger, A. Bondev, and R. Wetzker. 1999. Retinoic acid induces selective expression of phosphoinositide 3-kinase gamma in myelomonocytic U937 cells. Cell Growth Differ. 10:447-456. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

