Evolution of P elements in natural populations of Drosophila willistoni and D. sturtevanti
- PMID: 15579688
- PMCID: PMC1448778
- DOI: 10.1534/genetics.103.025775
Evolution of P elements in natural populations of Drosophila willistoni and D. sturtevanti
Abstract
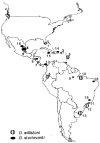
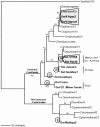
To determine how population structure of the host species affects the spread of transposable elements and to assess the strength of selection acting on different structural regions, we sequenced P elements from strains of Drosophila willistoni and Drosophila sturtevanti sampled from across the distributions of these species. Elements from D. sturtevanti exhibited considerable sequence variation, and similarity among them was correlated to geographic distance between collection sites. By contrast, all D. willistoni elements sampled were essentially identical (pi < 0.2%) and exhibited patterns typical of a recent population expansion. While the canonical P elements sampled from D. sturtevanti appear to be long-time residents in that species, a rapid expansion of a very young canonical P-element lineage is suggested in D. willistoni, overcoming barriers such as large geographical distances and moderate levels of population subdivision. Between-species comparisons reveal selective constraints on P-element evolution, as indicated by significantly different substitution rates in noncoding, silent, and replacement sites. Most remarkably, in addition to replacement sites, selection pressure appears to be strong in the first and third introns and in the 3' and 5' flanking regions.
Figures

Similar articles
-
P elements in the saltans group of Drosophila: a new evaluation of their distribution and number of genomic insertion sites.Mol Phylogenet Evol. 2004 Jul;32(1):383-7. doi: 10.1016/j.ympev.2004.01.005. Mol Phylogenet Evol. 2004. PMID: 15186822
-
Molecular evolution of P transposable elements in the genus Drosophila. I. The saltans and willistoni species groups.Mol Biol Evol. 1995 Sep;12(5):902-13. doi: 10.1093/oxfordjournals.molbev.a040267. Mol Biol Evol. 1995. PMID: 7476136
-
Harrow: new Drosophila hAT transposons involved in horizontal transfer.Insect Mol Biol. 2010 Apr;19(2):217-28. doi: 10.1111/j.1365-2583.2009.00977.x. Epub 2009 Dec 15. Insect Mol Biol. 2010. PMID: 20017754
-
Patterns of polymorphism and divergence from noncoding sequences of Drosophila melanogaster and D. simulans: evidence for nonequilibrium processes.Mol Biol Evol. 2005 Jan;22(1):51-62. doi: 10.1093/molbev/msh269. Epub 2004 Sep 29. Mol Biol Evol. 2005. PMID: 15456897 Review.
-
Why is the genome variable? Insights from Drosophila.Trends Genet. 1992 Oct;8(10):355-62. doi: 10.1016/0168-9525(92)90281-8. Trends Genet. 1992. PMID: 1335624 Review.
Cited by
-
CR1 clade of non-LTR retrotransposons from Maculinea butterflies (Lepidoptera: Lycaenidae): evidence for recent horizontal transmission.BMC Evol Biol. 2007 Jun 25;7:93. doi: 10.1186/1471-2148-7-93. BMC Evol Biol. 2007. PMID: 17588269 Free PMC article.
-
Contrasting patterns of transposable element insertions in Drosophila heat-shock promoters.PLoS One. 2009 Dec 29;4(12):e8486. doi: 10.1371/journal.pone.0008486. PLoS One. 2009. PMID: 20041194 Free PMC article.
-
Non-LTR retrotransposons in fungi.Funct Integr Genomics. 2009 Feb;9(1):27-42. doi: 10.1007/s10142-008-0093-8. Epub 2008 Aug 2. Funct Integr Genomics. 2009. PMID: 18677522
-
Transposon display supports transpositional activity of P elements in species of the saltans group of Drosophila.J Genet. 2007 Apr;86(1):37-43. doi: 10.1007/s12041-007-0005-z. J Genet. 2007. PMID: 17656847
-
Evidence of horizontal transfer of non-autonomous Lep1 Helitrons facilitated by host-parasite interactions.Sci Rep. 2014 May 30;4:5119. doi: 10.1038/srep05119. Sci Rep. 2014. PMID: 24874102 Free PMC article.
References
-
- Clark, J. B., T. K. Altheide, M. J. Schlosser and M. G. Kidwell, 1995. Molecular evolution of P transposable elements in the genus Drosophila. I. The saltans and willistoni species groups. Mol. Biol. Evol. 12: 902–913. - PubMed
-
- Clark, J. B., J. C. Silva and M. G. Kidwell, 2002 Evidence of horizontal transfer of P transposable elements, pp. 161–171 in Horizontal Gene Transfer, edited by M. Syvanen and C. I. Kado. Academic Press, San Diego.
-
- Engels, W., 1989 P elements in Drosophila melanogaster, pp. 437–483 in Mobile DNA, edited by D. E. Berg and M. M. Howe. American Society of Microbiology, Washington, DC.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

