Insertion of a classical nuclear import signal into the matrix domain of the Rous sarcoma virus Gag protein interferes with virus replication
- PMID: 15564464
- PMCID: PMC533892
- DOI: 10.1128/JVI.78.24.13534-13542.2004
Insertion of a classical nuclear import signal into the matrix domain of the Rous sarcoma virus Gag protein interferes with virus replication
Abstract
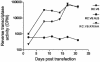
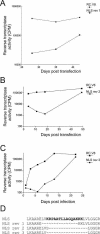
The Rous sarcoma virus Gag protein undergoes transient nuclear trafficking during virus assembly. Nuclear import is mediated by a nuclear targeting sequence within the MA domain. To gain insight into the role of nuclear transport in replication, we investigated whether addition of a "classical " nuclear localization signal (NLS) in Gag would affect virus assembly or infectivity. A bipartite NLS derived from nucleoplasmin was inserted into a region of the MA domain of Gag that is dispensable for budding and infectivity. Gag proteins bearing the nucleoplasmin NLS insertion displayed an assembly defect. Mutant virus particles (RC.V8.NLS) were not infectious, although they were indistinguishable from wild-type virions in Gag, Gag-Pol, Env, and genomic RNA incorporation and Gag protein processing. Unexpectedly, postinfection viral DNA synthesis was also normal, as similar amounts of two-long-terminal-repeat junction molecules were detected for RC.V8.NLS and wild type, suggesting that the replication block occurred after nuclear entry of proviral DNA. Phenotypically revertant viruses arose after continued passage in culture, and sequence analysis revealed that the nucleoplasmin NLS coding sequence was deleted from the gag gene. To determine whether the nuclear targeting activity of the nucleoplasmin sequence was responsible for the infectivity defect, two critical basic amino acids in the NLS were altered. This virus (RC.V8.KR/AA) had restored infectivity, and the MA.KR/AA protein showed reduced nuclear localization, comparable to the wild-type MA protein. These data demonstrate that addition of a second NLS, which might direct MA and/or Gag into the nucleus by an alternate import pathway, is not compatible with productive virus infection.
Figures


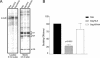
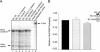

Similar articles
-
Genetic evidence for a connection between Rous sarcoma virus gag nuclear trafficking and genomic RNA packaging.J Virol. 2009 Jul;83(13):6790-7. doi: 10.1128/JVI.00101-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369339 Free PMC article.
-
Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein.Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):3944-9. doi: 10.1073/pnas.062652199. Epub 2002 Mar 12. Proc Natl Acad Sci U S A. 2002. PMID: 11891341 Free PMC article.
-
Overlapping roles of the Rous sarcoma virus Gag p10 domain in nuclear export and virion core morphology.J Virol. 2007 Oct;81(19):10718-28. doi: 10.1128/JVI.01061-07. Epub 2007 Jul 18. J Virol. 2007. PMID: 17634229 Free PMC article.
-
Bipartite nuclear localization sequence is indispensable for nuclear import and stability of self-dimerization of ADARa in Bombyx mori.Insect Biochem Mol Biol. 2024 Nov;174:104190. doi: 10.1016/j.ibmb.2024.104190. Epub 2024 Oct 9. Insect Biochem Mol Biol. 2024. PMID: 39389319 Review.
-
Beyond plasma membrane targeting: role of the MA domain of Gag in retroviral genome encapsidation.J Mol Biol. 2011 Jul 22;410(4):553-64. doi: 10.1016/j.jmb.2011.04.072. J Mol Biol. 2011. PMID: 21762800 Free PMC article. Review.
Cited by
-
Visualizing Rous Sarcoma Virus Genomic RNA Dimerization in the Nucleus, Cytoplasm, and at the Plasma Membrane.Viruses. 2021 May 13;13(5):903. doi: 10.3390/v13050903. Viruses. 2021. PMID: 34068261 Free PMC article.
-
The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles.J Virol. 2005 Nov;79(21):13463-72. doi: 10.1128/JVI.79.21.13463-13472.2005. J Virol. 2005. PMID: 16227267 Free PMC article.
-
Importin-beta family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid.J Virol. 2006 Feb;80(4):1798-806. doi: 10.1128/JVI.80.4.1798-1806.2006. J Virol. 2006. PMID: 16439536 Free PMC article.
-
Nuclear trafficking of retroviral RNAs and Gag proteins during late steps of replication.Viruses. 2013 Nov 18;5(11):2767-95. doi: 10.3390/v5112767. Viruses. 2013. PMID: 24253283 Free PMC article. Review.
-
Genetic evidence for a connection between Rous sarcoma virus gag nuclear trafficking and genomic RNA packaging.J Virol. 2009 Jul;83(13):6790-7. doi: 10.1128/JVI.00101-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369339 Free PMC article.
References
-
- Conti, E., and E. Izaurralde. 2001. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 13:310-319. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

