Genome structure and transcriptional regulation of human coronavirus NL63
- PMID: 15548333
- PMCID: PMC538260
- DOI: 10.1186/1743-422X-1-7
Genome structure and transcriptional regulation of human coronavirus NL63
Abstract
Background: Two human coronaviruses are known since the 1960s: HCoV-229E and HCoV-OC43. SARS-CoV was discovered in the early spring of 2003, followed by the identification of HCoV-NL63, the fourth member of the coronaviridae family that infects humans. In this study, we describe the genome structure and the transcription strategy of HCoV-NL63 by experimental analysis of the viral subgenomic mRNAs.
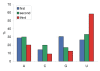
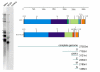
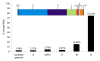
Results: The genome of HCoV-NL63 has the following gene order: 1a-1b-S-ORF3-E-M-N. The GC content of the HCoV-NL63 genome is extremely low (34%) compared to other coronaviruses, and we therefore performed additional analysis of the nucleotide composition. Overall, the RNA genome is very low in C and high in U, and this is also reflected in the codon usage. Inspection of the nucleotide composition along the genome indicates that the C-count increases significantly in the last one-third of the genome at the expense of U and G. We document the production of subgenomic (sg) mRNAs coding for the S, ORF3, E, M and N proteins. We did not detect any additional sg mRNA. Furthermore, we sequenced the 5' end of all sg mRNAs, confirming the presence of an identical leader sequence in each sg mRNA. Northern blot analysis indicated that the expression level among the sg mRNAs differs significantly, with the sg mRNA encoding nucleocapsid (N) being the most abundant.
Conclusions: The presented data give insight into the viral evolution and mutational patterns in coronaviral genome. Furthermore our data show that HCoV-NL63 employs the discontinuous replication strategy with generation of subgenomic mRNAs during the (-) strand synthesis. Because HCoV-NL63 has a low pathogenicity and is able to grow easily in cell culture, this virus can be a powerful tool to study SARS coronavirus pathogenesis.
Figures

Similar articles
-
Coronavirus HKU1 and other coronavirus infections in Hong Kong.J Clin Microbiol. 2006 Jun;44(6):2063-71. doi: 10.1128/JCM.02614-05. J Clin Microbiol. 2006. PMID: 16757599 Free PMC article.
-
Identification of a new human coronavirus.Nat Med. 2004 Apr;10(4):368-73. doi: 10.1038/nm1024. Epub 2004 Mar 21. Nat Med. 2004. PMID: 15034574 Free PMC article.
-
Mosaic structure of human coronavirus NL63, one thousand years of evolution.J Mol Biol. 2006 Dec 15;364(5):964-73. doi: 10.1016/j.jmb.2006.09.074. Epub 2006 Oct 3. J Mol Biol. 2006. PMID: 17054987 Free PMC article.
-
A new model for coronavirus transcription.Adv Exp Med Biol. 1998;440:215-9. doi: 10.1007/978-1-4615-5331-1_26. Adv Exp Med Biol. 1998. PMID: 9782283 Review.
-
Viral and Cellular mRNA Translation in Coronavirus-Infected Cells.Adv Virus Res. 2016;96:165-192. doi: 10.1016/bs.aivir.2016.08.001. Epub 2016 Sep 10. Adv Virus Res. 2016. PMID: 27712623 Free PMC article. Review.
Cited by
-
Crystallographic analysis of the N-terminal domain of Middle East respiratory syndrome coronavirus nucleocapsid protein.Acta Crystallogr F Struct Biol Commun. 2015 Aug;71(Pt 8):977-80. doi: 10.1107/S2053230X15010146. Epub 2015 Jul 28. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 26249685 Free PMC article.
-
Cytosine deamination and selection of CpG suppressed clones are the two major independent biological forces that shape codon usage bias in coronaviruses.Virology. 2007 Dec 20;369(2):431-42. doi: 10.1016/j.virol.2007.08.010. Epub 2007 Sep 19. Virology. 2007. PMID: 17881030 Free PMC article.
-
Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome.Virology. 2007 Oct 25;367(2):428-39. doi: 10.1016/j.virol.2007.06.009. Epub 2007 Jul 6. Virology. 2007. PMID: 17617433 Free PMC article.
-
The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2.Virology. 2007 Oct 25;367(2):367-74. doi: 10.1016/j.virol.2007.04.035. Epub 2007 Jul 12. Virology. 2007. PMID: 17631932 Free PMC article.
-
Crystallization and preliminary crystallographic study of Porcine epidemic diarrhea virus main protease in complex with an inhibitor.Acta Crystallogr F Struct Biol Commun. 2014 Dec 1;70(Pt 12):1608-11. doi: 10.1107/S2053230X14021876. Epub 2014 Nov 14. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 25484208 Free PMC article.
References
-
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. - DOI - PubMed
-
- Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, Ling AE, Chan PK, Tam JS, Zambon MC, Gopal R, Drosten C, van der Werf S, Escriou N, Manuguerra JC, Stohr K, Peiris JS, Osterhaus AD. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. - DOI - PMC - PubMed
-
- Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. - DOI - PMC - PubMed
-
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. - DOI - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

