The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B
- PMID: 15538381
- PMCID: PMC535089
- DOI: 10.1038/sj.emboj.7600477
The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B
Abstract
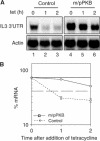
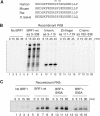
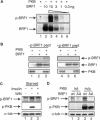
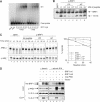
Butyrate response factor (BRF1) belongs to the Tis11 family of CCCH zinc-finger proteins, which bind to mRNAs containing an AU-rich element (ARE) in their 3' untranslated region and promote their deadenylation and rapid degradation. Independent signal transduction pathways have been reported to stabilize ARE-containing transcripts by a process thought to involve phosphorylation of ARE-binding proteins. Here we report that protein kinase B (PKB/Akt) stabilizes ARE transcripts by phosphorylating BRF1 at serine 92 (S92). Recombinant BRF1 promoted in vitro decay of ARE-containing mRNA (ARE-mRNA), yet phosphorylation by PKB impaired this activity. S92 phosphorylation of BRF1 did not impair ARE binding, but induced complex formation with the scaffold protein 14-3-3. In vivo and in vitro data support a model where PKB causes ARE-mRNA stabilization by inactivating BRF1 through binding to 14-3-3.
Figures


Similar articles
-
The AU-rich element mRNA decay-promoting activity of BRF1 is regulated by mitogen-activated protein kinase-activated protein kinase 2.RNA. 2008 May;14(5):950-9. doi: 10.1261/rna.983708. Epub 2008 Mar 6. RNA. 2008. PMID: 18326031 Free PMC article.
-
BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites.Mol Cell Biol. 2006 Dec;26(24):9497-507. doi: 10.1128/MCB.01099-06. Epub 2006 Oct 9. Mol Cell Biol. 2006. PMID: 17030608 Free PMC article.
-
The cAMP pathway regulates mRNA decay through phosphorylation of the RNA-binding protein TIS11b/BRF1.Mol Biol Cell. 2016 Dec 1;27(24):3841-3854. doi: 10.1091/mbc.E16-06-0379. Epub 2016 Oct 5. Mol Biol Cell. 2016. PMID: 27708140 Free PMC article.
-
The regulation and activities of the multifunctional serine/threonine kinase Akt/PKB.Exp Cell Res. 1999 Nov 25;253(1):210-29. doi: 10.1006/excr.1999.4690. Exp Cell Res. 1999. PMID: 10579924 Review.
-
Regulation of protein kinase B.J Recept Signal Transduct Res. 1999 Jan-Jul;19(1-4):121-8. doi: 10.3109/10799899909036639. J Recept Signal Transduct Res. 1999. PMID: 10071752 Review.
Cited by
-
RNA binding proteins as regulators of immune cell biology.Clin Exp Immunol. 2016 Jan;183(1):37-49. doi: 10.1111/cei.12684. Epub 2015 Sep 11. Clin Exp Immunol. 2016. PMID: 26201441 Free PMC article. Review.
-
Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):666-79. doi: 10.1016/j.bbagrm.2013.02.003. Epub 2013 Feb 18. Biochim Biophys Acta. 2013. PMID: 23428348 Free PMC article. Review.
-
CTGF facilitates cell-cell communication in chondrocytes via PI3K/Akt signalling pathway.Cell Prolif. 2021 Mar;54(3):e13001. doi: 10.1111/cpr.13001. Epub 2021 Feb 1. Cell Prolif. 2021. PMID: 33522639 Free PMC article.
-
Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-Regulation.Nucleic Acids Res. 2009 Jun;37(11):3612-24. doi: 10.1093/nar/gkp223. Epub 2009 Apr 9. Nucleic Acids Res. 2009. PMID: 19359363 Free PMC article.
-
The ribonome: a dominant force in co-ordinating gene expression.Biol Cell. 2009 Mar;101(3):169-81. doi: 10.1042/BC20080055. Biol Cell. 2009. PMID: 19152504 Free PMC article. Review.
References
-
- Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P (1996) Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett 399: 333–338 - PubMed
-
- Andjelkovic M, Alessi DR, Meier R, Fernandez A, Lamb NJ, Frech M, Cron P, Cohen P, Lucocq JM, Hemmings BA (1997) Role of translocation in the activation and function of protein kinase B. J Biol Chem 272: 31515–31524 - PubMed
-
- Briata P, Ilengo C, Corte G, Moroni C, Rosenfeld MG, Chen CY, Gherzi R (2003) The Wnt/beta-catenin → Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol Cell 12: 1201–1211 - PubMed
-
- Brook M, Sully G, Clark AR, Saklatvala J (2000) Regulation of tumour necrosis factor alpha mRNA stability by the mitogen-activated protein kinase p38 signalling cascade (in process citation). FEBS Lett 483: 57–61 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

