ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin
- PMID: 15507644
- PMCID: PMC525046
- DOI: 10.1128/JVI.78.22.12566-12575.2004
ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin
Abstract
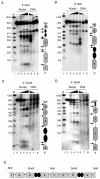
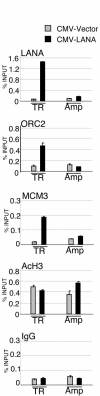
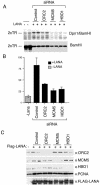
The viral genome of Kaposi's sarcoma-associated herpesvirus (KSHV) persists as an extrachromosomal plasmid in latently infected cells. The KSHV latency-associated nuclear antigen (LANA) stimulates plasmid maintenance and DNA replication by binding to an approximately 150-bp region within the viral terminal repeats (TR). We have used chromatin immunoprecipitation assays to demonstrate that LANA binds specifically to the replication origin sequence within the KSHV TR in latently infected cells. The latent replication origin within the TR was also bound by LANA-associated proteins CBP, double-bromodomain-containing protein 2 (BRD2), and the origin recognition complex 2 protein (ORC2) and was enriched in hyperacetylated histones H3 and H4 relative to other regions of the latent genome. Cell cycle analysis indicated that the minichromosome maintenance complex protein, MCM3, bound TR in late-G(1)/S-arrested cells, which coincided with the loss of histone H3 K4 methylation. Micrococcal nuclease studies revealed that TRs are embedded in a highly ordered nucleosome array that becomes disorganized in late G(1)/S phase. ORC binding to TR was LANA dependent when reconstituted in transfected plasmids. DNA affinity purification confirmed that LANA, CBP, BRD2, and ORC2 bound TR specifically and identified the histone acetyltransferase HBO1 (histone acetyltransferase binding to ORC1) as a potential TR binding protein. Disruption of ORC2, MCM5, and HBO1 expression by small interfering RNA reduced LANA-dependent DNA replication of TR-containing plasmids. These findings are the first demonstration that cellular replication and origin licensing factors are required for KSHV latent cycle replication. These results also suggest that the KSHV latent origin of replication is a unique chromatin environment containing histone H3 hyperacetylation within heterochromatic tandem repeats.
Figures


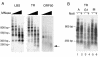

Similar articles
-
Minichromosome Maintenance Proteins Cooperate with LANA during the G1/S Phase of the Cell Cycle To Support Viral DNA Replication.J Virol. 2019 Mar 21;93(7):e02256-18. doi: 10.1128/JVI.02256-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651368 Free PMC article.
-
Latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats.J Virol. 2006 Mar;80(5):2243-56. doi: 10.1128/JVI.80.5.2243-2256.2006. J Virol. 2006. PMID: 16474132 Free PMC article.
-
The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells.J Virol. 2002 Nov;76(22):11677-87. doi: 10.1128/jvi.76.22.11677-11687.2002. J Virol. 2002. PMID: 12388727 Free PMC article.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
-
The latency-associated nuclear antigen, a multifunctional protein central to Kaposi's sarcoma-associated herpesvirus latency.Future Microbiol. 2011 Dec;6(12):1399-413. doi: 10.2217/fmb.11.137. Future Microbiol. 2011. PMID: 22122438 Free PMC article. Review.
Cited by
-
Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's Sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1.J Virol. 2005 Nov;79(21):13618-29. doi: 10.1128/JVI.79.21.13618-13629.2005. J Virol. 2005. PMID: 16227282 Free PMC article.
-
Complete in vitro reconstitution of adeno-associated virus DNA replication requires the minichromosome maintenance complex proteins.J Virol. 2008 Feb;82(3):1458-64. doi: 10.1128/JVI.01968-07. Epub 2007 Dec 5. J Virol. 2008. PMID: 18057257 Free PMC article.
-
Cell cycle-dependent chromatin shuttling of HBO1-JADE1 histone acetyl transferase (HAT) complex.Cell Cycle. 2014;13(12):1885-901. doi: 10.4161/cc.28759. Epub 2014 Apr 16. Cell Cycle. 2014. PMID: 24739512 Free PMC article.
-
Telomeres and viruses: common themes of genome maintenance.Front Oncol. 2012 Dec 31;2:201. doi: 10.3389/fonc.2012.00201. eCollection 2012. Front Oncol. 2012. PMID: 23293769 Free PMC article.
-
Timeless-dependent DNA replication-coupled recombination promotes Kaposi's Sarcoma-associated herpesvirus episome maintenance and terminal repeat stability.J Virol. 2013 Apr;87(7):3699-709. doi: 10.1128/JVI.02211-12. Epub 2013 Jan 16. J Virol. 2013. PMID: 23325691 Free PMC article.
References
-
- Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. - PubMed
-
- Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-asociated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. - PubMed
-
- Brewer, B. J., and W. L. Fangman. 1991. Mapping replication origins in yeast chromosomes. Bioessays 13:317-322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

