Cross talk between the +73/294 interaction and the cleavage site in RNase P RNA mediated cleavage
- PMID: 15477392
- PMCID: PMC524293
- DOI: 10.1093/nar/gkh883
Cross talk between the +73/294 interaction and the cleavage site in RNase P RNA mediated cleavage
Abstract
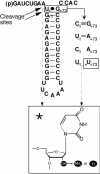
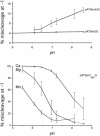
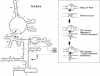
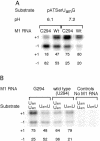
To monitor functionally important metal ions and possible cross talk in RNase P RNA mediated cleavage we studied cleavage of substrates, where the 2'OH at the RNase P cleavage site (at -1) and/or at position +73 had been replaced with a 2' amino group (or 2'H). Our data showed that the presence of 2' modifications at these positions affected cleavage site recognition, ground state binding of substrate and/or rate of cleavage. Cleavage of 2' amino substituted substrates at different pH showed that substitution of Mg2+ by Mn2+ (or Ca2+), identity of residues at and near the cleavage site, and addition of C5 protein influenced the frequency of miscleavage at -1 (cleavage at the correct site is referred to as +1). From this we infer that these findings point at effects mediated by protonation/deprotonation of the 2' amino group, i.e. an altered charge distribution, at the site of cleavage. Moreover, our data suggested that the structural architecture of the interaction between the 3' end of the substrate and RNase P RNA influence the charge distribution at the cleavage site as well as the rate of cleavage under conditions where the chemistry is suggested to be rate limiting. Thus, these data provide evidence for cross talk between the +73/294 interaction and the cleavage site in RNase P RNA mediated cleavage. We discuss the role metal ions might play in this cross talk and the likelihood that at least one functionally important metal ion is positioned in the vicinity of, and use the 2'OH at the cleavage site as an inner or outer sphere ligand.
Figures


Similar articles
-
Complexity in orchestration of chemical groups near different cleavage sites in RNase P RNA mediated cleavage.J Mol Biol. 2005 Aug 12;351(2):251-7. doi: 10.1016/j.jmb.2005.06.031. J Mol Biol. 2005. PMID: 16005891
-
The exocyclic amine at the RNase P cleavage site contributes to substrate binding and catalysis.J Mol Biol. 2006 Jun 9;359(3):572-84. doi: 10.1016/j.jmb.2006.03.040. Epub 2006 Apr 3. J Mol Biol. 2006. PMID: 16638615
-
The cleavage step of ribonuclease P catalysis is determined by ribozyme-substrate interactions both distal and proximal to the cleavage site.Biochemistry. 1999 Jul 6;38(27):8612-20. doi: 10.1021/bi990691f. Biochemistry. 1999. PMID: 10393536
-
Substrate discrimination in RNase P RNA-mediated cleavage: importance of the structural environment of the RNase P cleavage site.Nucleic Acids Res. 2005 Apr 7;33(6):2012-21. doi: 10.1093/nar/gki344. Print 2005. Nucleic Acids Res. 2005. PMID: 15817565 Free PMC article.
-
RNase P RNA mediated cleavage: substrate recognition and catalysis.Biochimie. 2007 Oct;89(10):1183-94. doi: 10.1016/j.biochi.2007.05.009. Epub 2007 Jun 2. Biochimie. 2007. PMID: 17624654 Review.
Cited by
-
Distributive enzyme binding controlled by local RNA context results in 3' to 5' directional processing of dicistronic tRNA precursors by Escherichia coli ribonuclease P.Nucleic Acids Res. 2019 Feb 20;47(3):1451-1467. doi: 10.1093/nar/gky1162. Nucleic Acids Res. 2019. PMID: 30496557 Free PMC article.
-
Nucleic acid catalysis: metals, nucleobases, and other cofactors.Chem Rev. 2014 Apr 23;114(8):4318-42. doi: 10.1021/cr400476k. Epub 2014 Apr 14. Chem Rev. 2014. PMID: 24730975 Free PMC article. Review. No abstract available.
-
A minimal RNA substrate with dual fluorescent probes enables rapid kinetics and provides insight into bacterial RNase P active site interactions.RSC Chem Biol. 2024 May 17;5(7):652-668. doi: 10.1039/d4cb00049h. eCollection 2024 Jul 3. RSC Chem Biol. 2024. PMID: 38966670 Free PMC article.
-
Cleavage of Model Substrates by Arabidopsis thaliana PRORP1 Reveals New Insights into Its Substrate Requirements.PLoS One. 2016 Aug 5;11(8):e0160246. doi: 10.1371/journal.pone.0160246. eCollection 2016. PLoS One. 2016. PMID: 27494328 Free PMC article.
-
The naturally trans-acting ribozyme RNase P RNA has leadzyme properties.Nucleic Acids Res. 2005 Dec 6;33(21):6920-30. doi: 10.1093/nar/gki993. Print 2005. Nucleic Acids Res. 2005. PMID: 16332695 Free PMC article.
References
-
- Doudna J.A. and Cech,T.R. (2002) The chemical repertoire of natural ribozymes. Nature, 418, 222–228. - PubMed
-
- Guerrier-Takada C., Gardiner,K., Marsh,T., Pace,N. and Altman,S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell, 35, 849–857. - PubMed
-
- Altman S. and Kirsebom,L.A. (1999) Ribonuclease,P. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds.), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 351–380.
-
- Vioque A., Arnez,J. and Altman,S. (1988) Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J. Mol. Biol., 202, 835–848. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

