Distinct in vivo dynamics of vertebrate SUMO paralogues
- PMID: 15456902
- PMCID: PMC532004
- DOI: 10.1091/mbc.e04-07-0589
Distinct in vivo dynamics of vertebrate SUMO paralogues
Abstract
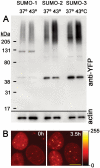
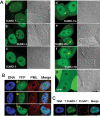
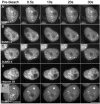
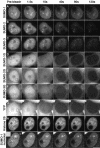
There are three mammalian SUMO paralogues: SUMO-1 is approximately 45% identical to SUMO-2 and SUMO-3, which are 96% identical to each other. It is currently unclear whether SUMO-1, -2, and -3 function in ways that are unique, redundant, or antagonistic. To address this question, we examined the dynamics of individual SUMO paralogues by using cell lines that stably express each of the mammalian SUMO proteins fused to the yellow fluorescent protein (YFP). Whereas SUMO-2 and -3 showed very similar distributions throughout the nucleoplasm, SUMO-1 was uniquely distributed to the nuclear envelope and to the nucleolus. Photobleaching experiments revealed that SUMO-1 dynamics was much slower than SUMO-2 and -3 dynamics. Additionally, the mobility of SUMO paralogues differed between subnuclear structures. Finally, the timing and distributions were dissimilar between paralogues as cells exited from mitosis. SUMO-1 was recruited to nuclear membrane as nuclear envelopes reformed in late anaphase, and accumulated rapidly into the nucleus. SUMO-2 and SUMO-3 localized to chromosome earlier and accumulated gradually during telophase. Together, these findings demonstrate that mammalian SUMO-1 shows patterns of utilization that are clearly discrete from the patterns of SUMO-2 and -3 throughout the cell cycle, arguing that it is functionally distinct and specifically regulated in vivo.
Figures



Similar articles
-
SUMO paralogue-specific functions revealed through systematic analysis of human knockout cell lines and gene expression data.Mol Biol Cell. 2021 Sep 1;32(19):1849-1866. doi: 10.1091/mbc.E21-01-0031. Epub 2021 Jul 7. Mol Biol Cell. 2021. PMID: 34232706 Free PMC article.
-
Molecular features of human ubiquitin-like SUMO genes and their encoded proteins.Gene. 2002 Aug 21;296(1-2):65-73. doi: 10.1016/s0378-1119(02)00843-0. Gene. 2002. PMID: 12383504
-
SUMOylation of hypoxia-inducible factor-1alpha reduces its transcriptional activity.Biochem Biophys Res Commun. 2007 Aug 31;360(3):646-52. doi: 10.1016/j.bbrc.2007.06.103. Epub 2007 Jun 27. Biochem Biophys Res Commun. 2007. PMID: 17610843
-
Strategies for the expression of SUMO-modified target proteins in Escherichia coli.Methods Mol Biol. 2009;497:211-21. doi: 10.1007/978-1-59745-566-4_14. Methods Mol Biol. 2009. PMID: 19107420 Review.
-
Enhanced detection of in vivo SUMO conjugation by Ubc9 fusion-dependent sumoylation (UFDS).Methods Mol Biol. 2009;497:63-79. doi: 10.1007/978-1-59745-566-4_5. Methods Mol Biol. 2009. PMID: 19107411 Review.
Cited by
-
PML body meets telomere: the beginning of an ALTernate ending?Nucleus. 2012 May-Jun;3(3):263-75. doi: 10.4161/nucl.20326. Epub 2012 May 1. Nucleus. 2012. PMID: 22572954 Free PMC article. Review.
-
SUSP1 antagonizes formation of highly SUMO2/3-conjugated species.J Cell Biol. 2006 Sep 25;174(7):939-49. doi: 10.1083/jcb.200510103. J Cell Biol. 2006. PMID: 17000875 Free PMC article.
-
Age Alters Chromatin Structure and Expression of SUMO Proteins under Stress Conditions in Human Adipose-Derived Stem Cells.Sci Rep. 2018 Jul 31;8(1):11502. doi: 10.1038/s41598-018-29775-y. Sci Rep. 2018. PMID: 30065345 Free PMC article.
-
The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing.EMBO Rep. 2008 Mar;9(3):273-9. doi: 10.1038/embor.2008.3. Epub 2008 Feb 8. EMBO Rep. 2008. PMID: 18259216 Free PMC article.
-
Defective sumoylation pathway directs congenital heart disease.Birth Defects Res A Clin Mol Teratol. 2011 Jun;91(6):468-76. doi: 10.1002/bdra.20816. Epub 2011 May 11. Birth Defects Res A Clin Mol Teratol. 2011. PMID: 21563299 Free PMC article.
References
-
- Bachant, J., Alcasabas, A., Blat, Y., Kleckner, N., and Elledge, S.J. (2002). The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9, 1169–1182. - PubMed
-
- Dieckhoff, P., Bolte, M., Sancak, Y., Braus, G.H., and Irniger, S. (2004). Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol. Microbiol. 51, 1375–1387. - PubMed
-
- Joseph, J., Liu, S. T., Jablonski, S. A., Yen, T. J., Dosso, M. (2004). The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14, 611–617. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

