Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to Its native promoter and the defense-related gene PcPR1-1 in Parsley
- PMID: 15367720
- PMCID: PMC520956
- DOI: 10.1105/tpc.104.024810
Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to Its native promoter and the defense-related gene PcPR1-1 in Parsley
Abstract
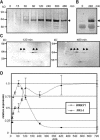
WRKY transcription factors form a large family that plays a role in plant responses to biotic stress and during senescence. Defining in vivo relevant WRKY/promoter relationships has been hampered by the factors' indiscriminate binding to known W box DNA elements and their possible genetic redundance. Employing chromatin immunoprecipitations (ChIP) of cultured cells, we show that parsley (Petroselinum crispum) WRKY1 protein binds to the W boxes of its native promoter as well as to that of PcWRKY3 and the defense-related PR10-class marker gene Pathogenesis-Related1-1 (PcPR1-1). Although present at low concentrations in resting cells, WRKY1 does not appear to play a role in the immediate early gene response upon elicitation because it does not bind to the promoter at this time. Paradoxically, in vivo binding at the PcWRKY1 promoter correlates more with downregulation of gene expression, whereas previous overexpression studies suggested an activating function of WRKY1 on PcWRKY1 expression. By contrast, PcPR1-1 expression remains strong when its promoter is occupied in vivo by WRKY1. Unexpectedly, ChIP revealed that W boxes at promoter sites are constitutively occupied by other WRKY transcription factors, indicating that site recruitment does not seem to play a major role in their regulation. Rather, WRKY proteins very likely act in a network of mutually competing participants with temporal displacement occurring at defined preoccupied sites by other family members in a stimulus-dependent manner.
Figures

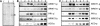



Similar articles
-
Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley.Biochim Biophys Acta. 2002 Jun 7;1576(1-2):92-100. doi: 10.1016/s0167-4781(02)00298-1. Biochim Biophys Acta. 2002. PMID: 12031488
-
Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors.EMBO J. 1999 Sep 1;18(17):4689-99. doi: 10.1093/emboj/18.17.4689. EMBO J. 1999. PMID: 10469648 Free PMC article.
-
Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes.EMBO J. 1996 Oct 15;15(20):5690-700. EMBO J. 1996. PMID: 8896462 Free PMC article.
-
Protein-protein interactions in the regulation of WRKY transcription factors.Mol Plant. 2013 Mar;6(2):287-300. doi: 10.1093/mp/sst026. Epub 2013 Mar 2. Mol Plant. 2013. PMID: 23455420 Review.
-
Regulatory elements required for light-mediated expression of the Petroselinum crispum chalcone synthase gene.Symp Soc Exp Biol. 1991;45:191-210. Symp Soc Exp Biol. 1991. PMID: 1843408 Review.
Cited by
-
Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene.Plant Mol Biol. 2007 Jun;64(3):293-303. doi: 10.1007/s11103-007-9152-0. Epub 2007 Mar 28. Plant Mol Biol. 2007. PMID: 17390108
-
Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP).Nat Protoc. 2010 Mar;5(3):457-72. doi: 10.1038/nprot.2009.244. Epub 2010 Feb 18. Nat Protoc. 2010. PMID: 20203663
-
Role of PsnWRKY70 in Regulatory Network Response to Infection with Alternaria alternata (Fr.) Keissl in Populus.Int J Mol Sci. 2022 Jul 7;23(14):7537. doi: 10.3390/ijms23147537. Int J Mol Sci. 2022. PMID: 35886886 Free PMC article.
-
Requirement of TCTG(G/C) Direct Repeats and Overlapping GATA Site for Maintaining the Cardiac-Specific Expression of Cardiac troponin T in Developing and Adult Mice.Anat Rec (Hoboken). 2008 Dec;291(12):1574-86. doi: 10.1002/ar.20772. Anat Rec (Hoboken). 2008. PMID: 18951515 Free PMC article.
-
Overexpression of ZmWRKY65 transcription factor from maize confers stress resistances in transgenic Arabidopsis.Sci Rep. 2021 Feb 17;11(1):4024. doi: 10.1038/s41598-021-83440-5. Sci Rep. 2021. PMID: 33597656 Free PMC article.
References
-
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. - PubMed
-
- Barolo, S., and Posakony, J.W. (2002). Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167–1181. - PubMed
-
- Cahill, M.A., Ernst, W.H., Janknecht, R., and Nordheim, A. (1994). Regulatory squelching. FEBS Lett. 344, 105–108. - PubMed
-
- Chen, C., and Chen, Z. (2000). Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol. Biol. 42, 387–396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

