Single nucleotide polymorphism-based validation of exonic splicing enhancers
- PMID: 15340491
- PMCID: PMC514884
- DOI: 10.1371/journal.pbio.0020268
Single nucleotide polymorphism-based validation of exonic splicing enhancers
Abstract
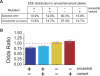
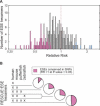
Because deleterious alleles arising from mutation are filtered by natural selection, mutations that create such alleles will be underrepresented in the set of common genetic variation existing in a population at any given time. Here, we describe an approach based on this idea called VERIFY (variant elimination reinforces functionality), which can be used to assess the extent of natural selection acting on an oligonucleotide motif or set of motifs predicted to have biological activity. As an application of this approach, we analyzed a set of 238 hexanucleotides previously predicted to have exonic splicing enhancer (ESE) activity in human exons using the relative enhancer and silencer classification by unanimous enrichment (RESCUE)-ESE method. Aligning the single nucleotide polymorphisms (SNPs) from the public human SNP database to the chimpanzee genome allowed inference of the direction of the mutations that created present-day SNPs. Analyzing the set of SNPs that overlap RESCUE-ESE hexamers, we conclude that nearly one-fifth of the mutations that disrupt predicted ESEs have been eliminated by natural selection (odds ratio = 0.82 +/- 0.05). This selection is strongest for the predicted ESEs that are located near splice sites. Our results demonstrate a novel approach for quantifying the extent of natural selection acting on candidate functional motifs and also suggest certain features of mutations/SNPs, such as proximity to the splice site and disruption or alteration of predicted ESEs, that should be useful in identifying variants that might cause a biological phenotype.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures


Similar articles
-
Synonymous SNPs provide evidence for selective constraint on human exonic splicing enhancers.J Mol Evol. 2006 Jan;62(1):89-98. doi: 10.1007/s00239-005-0055-x. Epub 2005 Nov 30. J Mol Evol. 2006. PMID: 16320116
-
Missense mutations in cancer suppressor gene TP53 are colocalized with exonic splicing enhancers (ESEs).Mutat Res. 2004 Oct 4;554(1-2):175-83. doi: 10.1016/j.mrfmmm.2004.04.014. Mutat Res. 2004. PMID: 15450416
-
Why Selection Might Be Stronger When Populations Are Small: Intron Size and Density Predict within and between-Species Usage of Exonic Splice Associated cis-Motifs.Mol Biol Evol. 2015 Jul;32(7):1847-61. doi: 10.1093/molbev/msv069. Epub 2015 Mar 13. Mol Biol Evol. 2015. PMID: 25771198 Free PMC article.
-
Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases.Trends Biochem Sci. 2000 Mar;25(3):106-10. doi: 10.1016/s0968-0004(00)01549-8. Trends Biochem Sci. 2000. PMID: 10694877 Review.
-
[Pathogenic mutation or polymorphism? (How to find criteria)].Med Wieku Rozwoj. 2006 Jan-Mar;10(1 Pt 2):335-41. Med Wieku Rozwoj. 2006. PMID: 17028397 Review. Polish.
Cited by
-
Testing for natural selection in human exonic splicing regulators associated with evolutionary rate shifts.J Mol Evol. 2013 Apr;76(4):228-39. doi: 10.1007/s00239-013-9555-2. Epub 2013 Mar 26. J Mol Evol. 2013. PMID: 23529588
-
Splicing of long non-coding RNAs primarily depends on polypyrimidine tract and 5' splice-site sequences due to weak interactions with SR proteins.Nucleic Acids Res. 2019 Jan 25;47(2):911-928. doi: 10.1093/nar/gky1147. Nucleic Acids Res. 2019. PMID: 30445574 Free PMC article.
-
Functional single nucleotide polymorphism-based association studies.Hum Genomics. 2006 Jun;2(6):391-402. doi: 10.1186/1479-7364-2-6-391. Hum Genomics. 2006. PMID: 16848977 Free PMC article. Review.
-
Genomic selective constraints in murid noncoding DNA.PLoS Genet. 2006 Nov 24;2(11):e204. doi: 10.1371/journal.pgen.0020204. Epub 2006 Oct 18. PLoS Genet. 2006. PMID: 17166057 Free PMC article.
-
Alternative approach to a heavy weight problem.Genome Res. 2008 Feb;18(2):214-20. doi: 10.1101/gr.6661308. Epub 2007 Dec 20. Genome Res. 2008. PMID: 18096750 Free PMC article.
References
-
- Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. - PubMed
-
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. - PubMed
-
- Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, et al. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11:1005–1011. - PubMed
-
- Blencowe BJ. Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

