cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four
- PMID: 15340080
- PMCID: PMC515050
- DOI: 10.1128/MCB.24.18.8210-8220.2004
cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four
Abstract
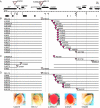
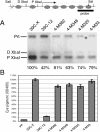
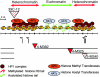
The heterochromatic domains of Drosophila melanogaster (pericentric heterochromatin, telomeres, and the fourth chromosome) are characterized by histone hypoacetylation, high levels of histone H3 methylated on lysine 9 (H3-mK9), and association with heterochromatin protein 1 (HP1). While the specific interaction of HP1 with both H3-mK9 and histone methyltransferases suggests a mechanism for the maintenance of heterochromatin, it leaves open the question of how heterochromatin formation is targeted to specific domains. Expression characteristics of reporter transgenes inserted at different sites in the fourth chromosome define a minimum of three euchromatic and three heterochromatic domains, interspersed. Here we searched for cis-acting DNA sequence determinants that specify heterochromatic domains. Genetic screens for a switch in phenotype demonstrate that local deletions or duplications of 5 to 80 kb of DNA flanking a transposon reporter can lead to the loss or acquisition of variegation, pointing to short-range cis-acting determinants for silencing. This silencing is dependent on HP1. A switch in transgene expression correlates with a switch in chromatin structure, judged by nuclease accessibility. Mapping data implicate the 1360 transposon as a target for heterochromatin formation. We propose that heterochromatin formation is initiated at dispersed repetitive elements along the fourth chromosome and spreads for approximately 10 kb or until encountering competition from a euchromatic determinant.
Figures
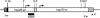
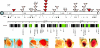


Similar articles
-
An investigation of heterochromatin domains on the fourth chromosome of Drosophila melanogaster.Genetics. 2008 Mar;178(3):1177-91. doi: 10.1534/genetics.107.081828. Epub 2008 Feb 1. Genetics. 2008. PMID: 18245350 Free PMC article.
-
Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing.EMBO J. 2002 Mar 1;21(5):1121-31. doi: 10.1093/emboj/21.5.1121. EMBO J. 2002. PMID: 11867540 Free PMC article.
-
Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery.Science. 2004 Jan 30;303(5658):669-72. doi: 10.1126/science.1092653. Science. 2004. PMID: 14752161
-
Does heterochromatin protein 1 always follow code?Proc Natl Acad Sci U S A. 2002 Dec 10;99 Suppl 4(Suppl 4):16462-9. doi: 10.1073/pnas.162371699. Epub 2002 Jul 31. Proc Natl Acad Sci U S A. 2002. PMID: 12151603 Free PMC article. Review.
-
Intercalary heterochromatin in polytene chromosomes of Drosophila melanogaster.Chromosoma. 2008 Oct;117(5):411-8. doi: 10.1007/s00412-008-0163-7. Epub 2008 May 20. Chromosoma. 2008. PMID: 18491121 Review.
Cited by
-
POF regulates the expression of genes on the fourth chromosome in Drosophila melanogaster by binding to nascent RNA.Mol Cell Biol. 2012 Jun;32(11):2121-34. doi: 10.1128/MCB.06622-11. Epub 2012 Apr 2. Mol Cell Biol. 2012. PMID: 22473994 Free PMC article.
-
High-resolution mapping reveals links of HP1 with active and inactive chromatin components.PLoS Genet. 2007 Mar 2;3(3):e38. doi: 10.1371/journal.pgen.0030038. PLoS Genet. 2007. PMID: 17335352 Free PMC article.
-
Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements.Proc Natl Acad Sci U S A. 2012 Aug 28;109(35):14104-9. doi: 10.1073/pnas.1207036109. Epub 2012 Aug 13. Proc Natl Acad Sci U S A. 2012. PMID: 22891327 Free PMC article.
-
Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila.Cold Spring Harb Perspect Biol. 2013 Aug 1;5(8):a017780. doi: 10.1101/cshperspect.a017780. Cold Spring Harb Perspect Biol. 2013. PMID: 23906716 Free PMC article. Review.
-
Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA.PLoS Genet. 2012 Feb;8(2):e1002474. doi: 10.1371/journal.pgen.1002474. Epub 2012 Feb 9. PLoS Genet. 2012. PMID: 22346759 Free PMC article.
References
-
- Adams, M. D., and J. J. Sekelsky. 2002. From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nat. Rev. Genet. 3:189-198. - PubMed
-
- Ahmad, K., and S. Henikoff. 2001. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104:839-847. - PubMed
-
- Aravin, A. A., N. A. Naumova, A. V. Tulin, V. V. Vagin, Y. M. Rozosky, and V. A. Gvozdev. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11:1017-1027. - PubMed
-
- Aravin, A. A., M. Lagos-Quintana, A. Yalcin, M. Zavolan, D. Marks, B. Snyder, T. Gaasterland, J. Meyer, and T. Tuschl. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337-350. - PubMed
-
- Barigozzi, C., S. Dolfini, M. Fracacaro, G. Rezzonico-Raimondi, and L. Tiepolo. 1966. In vitro study of the DNA replication patterns of somatic chromosomes of Drosophila melanogaster. Exp. Cell Res. 43:231-234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
