A novel type of silencing factor, Clr2, is necessary for transcriptional silencing at various chromosomal locations in the fission yeast Schizosaccharomyces pombe
- PMID: 15317867
- PMCID: PMC516054
- DOI: 10.1093/nar/gkh780
A novel type of silencing factor, Clr2, is necessary for transcriptional silencing at various chromosomal locations in the fission yeast Schizosaccharomyces pombe
Abstract
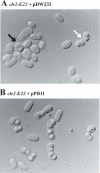
The mating-type region of the fission yeast Schizosaccharomyces pombe comprises three loci: mat1, mat2-P and mat3-M. mat1 is expressed and determines the mating type of the cell. mat2-P and mat3-M are two storage cassettes located in a 17 kb heterochromatic region with features identical to those of mammalian heterochromatin. Mutations in the swi6+, clr1+, clr2+, clr3+, clr4+ and clr6+ genes were obtained in screens for factors necessary for silencing the mat2-P-mat3-M region. swi6+ encodes a chromodomain protein, clr3+ and clr6+ histone deacetylases, and clr4+ a histone methyltransferase. Here, we describe the cloning and characterization of clr2+. The clr2+ gene encodes a 62 kDa protein with no obvious sequence homologs. Deletion of clr2+ not only affects transcriptional repression in the mating-type region, but also centromeric silencing and silencing of a PolII-transcribed gene inserted in the rDNA repeats. Using chromatin immunoprecipitation, we show that Clr2 is necessary for histone hypoacetylation in the mating-type region, suggesting that Clr2 acts upstream of histone deacetylases to promote transcriptional silencing.
Figures

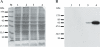

Similar articles
-
Transcriptional silencing in fission yeast.J Cell Physiol. 2000 Sep;184(3):311-8. doi: 10.1002/1097-4652(200009)184:3<311::AID-JCP4>3.0.CO;2-D. J Cell Physiol. 2000. PMID: 10911361 Review.
-
Silencing motifs in the Clr2 protein from fission yeast, Schizosaccharomyces pombe.PLoS One. 2014 Jan 27;9(1):e86948. doi: 10.1371/journal.pone.0086948. eCollection 2014. PLoS One. 2014. PMID: 24475199 Free PMC article.
-
Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination.Cell. 2004 Nov 12;119(4):469-80. doi: 10.1016/j.cell.2004.10.020. Cell. 2004. PMID: 15537537
-
New insights into donor directionality of mating-type switching in Schizosaccharomyces pombe.PLoS Genet. 2018 May 31;14(5):e1007424. doi: 10.1371/journal.pgen.1007424. eCollection 2018 May. PLoS Genet. 2018. PMID: 29852001 Free PMC article.
-
Studies on the mechanism of RNAi-dependent heterochromatin assembly.Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review.
Cited by
-
Intricate regulation on epigenetic stability of the subtelomeric heterochromatin and the centromeric chromatin in fission yeast.Curr Genet. 2019 Apr;65(2):381-386. doi: 10.1007/s00294-018-0886-9. Epub 2018 Sep 22. Curr Genet. 2019. PMID: 30244281 Review.
-
Structural insights into the binding mechanism of Clr4 methyltransferase to H3K9 methylated nucleosome.Sci Rep. 2024 Mar 5;14(1):5438. doi: 10.1038/s41598-024-56248-2. Sci Rep. 2024. PMID: 38443490 Free PMC article.
-
SHREC Silences Heterochromatin via Distinct Remodeling and Deacetylation Modules.Mol Cell. 2016 Apr 21;62(2):207-221. doi: 10.1016/j.molcel.2016.03.016. Mol Cell. 2016. PMID: 27105116 Free PMC article.
-
Advancing our understanding of functional genome organisation through studies in the fission yeast.Curr Genet. 2011 Feb;57(1):1-12. doi: 10.1007/s00294-010-0327-x. Epub 2010 Nov 27. Curr Genet. 2011. PMID: 21113595 Free PMC article. Review.
-
HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms.Mol Cell. 2008 Dec 26;32(6):778-90. doi: 10.1016/j.molcel.2008.10.026. Mol Cell. 2008. PMID: 19111658 Free PMC article.
References
-
- Arcangioli B. and Thon,G. (2004) Mating-type cassettes: structure, switching and silencing. In Egel,R. (ed.), The Molecular Biology of Schizosaccharomyces pombe. Springer, New York, NY, pp. 129–148.
-
- Egel R., Willer,M. and Nielsen,O. (1989) Unblocking of meiotic crossing-over between the silent mating-type cassettes of fission yeast, conditioned by the pleiotropic mutant rik1. Curr. Genet., 15, 407–410.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

