UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant
- PMID: 15314026
- PMCID: PMC514181
- DOI: 10.1101/gad.1219104
UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant
Abstract
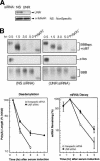
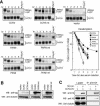
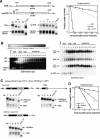
Messenger RNA decay mediated by the c-fos major protein coding-region determinant of instability (mCRD) is a useful system for studying translationally coupled mRNA turnover. Among the five mCRD-associated proteins identified previously, UNR was found to be an mCRD-binding protein and also a PABP-interacting protein. Interaction between UNR and PABP is necessary for the full destabilization function of the mCRD. By testing different classes of mammalian poly(A) nucleases, we identified CCR4 as a poly(A) nuclease involved in the mCRD-mediated rapid deadenylation in vivo and also associated with UNR. Blocking either translation initiation or elongation greatly impeded poly(A) shortening and mRNA decay mediated by the mCRD, demonstrating that the deadenylation step is coupled to ongoing translation of the message. These findings suggest a model in which the mCRD/UNR complex serves as a "landing/assembly" platform for formation of a deadenylation/decay mRNA-protein complex on an mCRD-containing transcript. The complex is dormant prior to translation. Accelerated deadenylation and decay of the transcript follows ribosome transit through the mCRD. This study provides new insights into a mechanism by which interplay between mRNA turnover and translation determines the lifespan of an mCRD-containing mRNA in the cytoplasm.
Figures




Similar articles
-
A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex.Cell. 2000 Sep 29;103(1):29-40. doi: 10.1016/s0092-8674(00)00102-1. Cell. 2000. PMID: 11051545
-
Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation.Mol Cell. 2009 Sep 24;35(6):868-80. doi: 10.1016/j.molcel.2009.08.004. Epub 2009 Aug 27. Mol Cell. 2009. PMID: 19716330 Free PMC article.
-
PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay.Mol Cell. 2018 Jun 21;70(6):1081-1088.e5. doi: 10.1016/j.molcel.2018.05.009. Mol Cell. 2018. PMID: 29932901
-
Poly(A)-binding proteins: structure, domain organization, and activity regulation.Biochemistry (Mosc). 2013 Dec;78(13):1377-91. doi: 10.1134/S0006297913130014. Biochemistry (Mosc). 2013. PMID: 24490729 Review.
-
Regulation of poly(A)-binding protein through PABP-interacting proteins.Cold Spring Harb Symp Quant Biol. 2006;71:537-43. doi: 10.1101/sqb.2006.71.061. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381337 Review.
Cited by
-
The DDX6-4E-T interaction mediates translational repression and P-body assembly.Nucleic Acids Res. 2016 Jul 27;44(13):6318-34. doi: 10.1093/nar/gkw565. Epub 2016 Jun 24. Nucleic Acids Res. 2016. PMID: 27342281 Free PMC article.
-
Cold shock domain-containing protein E1 is a posttranscriptional regulator of the LDL receptor.Sci Transl Med. 2022 Sep 14;14(662):eabj8670. doi: 10.1126/scitranslmed.abj8670. Epub 2022 Sep 14. Sci Transl Med. 2022. PMID: 36103516 Free PMC article.
-
Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation.Mol Cell Biol. 2007 Nov;27(22):7791-801. doi: 10.1128/MCB.01254-07. Epub 2007 Sep 4. Mol Cell Biol. 2007. PMID: 17785442 Free PMC article.
-
Coding region: the neglected post-transcriptional code.RNA Biol. 2011 Jan-Feb;8(1):44-8. doi: 10.4161/rna.8.1.13863. Epub 2011 Jan 1. RNA Biol. 2011. PMID: 21289484 Free PMC article. Review.
-
Mechanisms of deadenylation-dependent decay.Wiley Interdiscip Rev RNA. 2011 Mar-Apr;2(2):167-83. doi: 10.1002/wrna.40. Epub 2010 Sep 15. Wiley Interdiscip Rev RNA. 2011. PMID: 21957004 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
