Proteome reference maps of vegetative tissues in pea. An investigation of nitrogen mobilization from leaves during seed filling
- PMID: 15299134
- PMCID: PMC520794
- DOI: 10.1104/pp.104.041947
Proteome reference maps of vegetative tissues in pea. An investigation of nitrogen mobilization from leaves during seed filling
Abstract
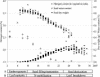
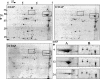
A proteomic approach was used to analyze protein changes during nitrogen mobilization (N mobilization) from leaves to filling seeds in pea (Pisum sativum). First, proteome reference maps were established for mature leaves and stems. They displayed around 190 Coomassie Blue-stained spots with pIs from 4 to 7. A total of 130 spots were identified by mass spectrometry as corresponding to 80 different proteins implicated in a variety of cellular functions. Although the leaf proteome map contained more abundant spots, corresponding to proteins involved in energy/carbon metabolism, than the stem map, their comparison revealed a highly similar protein profile. Second, the leaf proteome map was used to analyze quantitative variations in leaf proteins during N mobilization. Forty percent of the spots showed significant changes in their relative abundance in the total protein extract. The results confirmed the importance of Rubisco as a source of mobilizable nitrogen, and suggested that in pea leaves the rate of degradation of Rubisco may vary throughout N mobilization. Correlated with the loss of Rubisco was an increase in relative abundance of chloroplastic protease regulatory subunits. Concomitantly, the relative abundance of some proteins related to the photosynthetic apparatus (Rubisco activase, Rubisco-binding proteins) and of several chaperones increased. A role for these proteins in the maintenance of a Rubisco activation state and in the PSII repair during the intense proteolytic activity within the chloroplasts was proposed. Finally, two 14-3-3-like proteins, with a potential regulatory role, displayed differential expression patterns during the massive remobilization of nitrogen.
Figures


Similar articles
-
Dynamics of exogenous nitrogen partitioning and nitrogen remobilization from vegetative organs in pea revealed by 15N in vivo labeling throughout seed filling.Plant Physiol. 2005 Apr;137(4):1463-73. doi: 10.1104/pp.104.056713. Epub 2005 Mar 25. Plant Physiol. 2005. PMID: 15793068 Free PMC article.
-
Dissecting the proteome of pea mature seeds reveals the phenotypic plasticity of seed protein composition.Proteomics. 2009 Jan;9(2):254-71. doi: 10.1002/pmic.200700903. Proteomics. 2009. PMID: 19086096
-
Improvement of pea biomass and seed productivity by simultaneous increase of phloem and embryo loading with amino acids.Plant J. 2015 Jan;81(1):134-46. doi: 10.1111/tpj.12716. Epub 2014 Dec 3. Plant J. 2015. PMID: 25353986
-
The contrasting N management of two oilseed rape genotypes reveals the mechanisms of proteolysis associated with leaf N remobilization and the respective contributions of leaves and stems to N storage and remobilization during seed filling.BMC Plant Biol. 2015 Feb 21;15:59. doi: 10.1186/s12870-015-0437-1. BMC Plant Biol. 2015. PMID: 25848818 Free PMC article.
-
Manipulation of sucrose phloem and embryo loading affects pea leaf metabolism, carbon and nitrogen partitioning to sinks as well as seed storage pools.Plant J. 2020 Jan;101(1):217-236. doi: 10.1111/tpj.14533. Epub 2019 Sep 14. Plant J. 2020. PMID: 31520495
Cited by
-
Dynamics of exogenous nitrogen partitioning and nitrogen remobilization from vegetative organs in pea revealed by 15N in vivo labeling throughout seed filling.Plant Physiol. 2005 Apr;137(4):1463-73. doi: 10.1104/pp.104.056713. Epub 2005 Mar 25. Plant Physiol. 2005. PMID: 15793068 Free PMC article.
-
Soybean 14-3-3 gene family: identification and molecular characterization.Planta. 2011 Mar;233(3):569-82. doi: 10.1007/s00425-010-1315-6. Epub 2010 Nov 26. Planta. 2011. PMID: 21120521
-
Towards systems biological understanding of leaf senescence.Plant Mol Biol. 2013 Aug;82(6):519-28. doi: 10.1007/s11103-012-9974-2. Epub 2012 Oct 13. Plant Mol Biol. 2013. PMID: 23065109 Review.
-
Proteome Analysis of Poplar Seed Vigor.PLoS One. 2015 Jul 14;10(7):e0132509. doi: 10.1371/journal.pone.0132509. eCollection 2015. PLoS One. 2015. PMID: 26172265 Free PMC article.
-
Emerging Genomic Tools for Legume Breeding: Current Status and Future Prospects.Front Plant Sci. 2016 May 2;7:455. doi: 10.3389/fpls.2016.00455. eCollection 2016. Front Plant Sci. 2016. PMID: 27199998 Free PMC article. Review.
References
-
- Adam Z, Clarke AK (2002) Cutting edge of chloroplast proteolysis. Trends Plant Sci 7: 451–456 - PubMed
-
- Bantignies B, Seguin I, Muzac I, Dedaldechamp F, Gulick P, Ibrahim R (2000) Direct evidence for ribonucleolytic activity of a PR-10-like protein from white lupin roots. Plant Mol Biol 42: 871–881 - PubMed
-
- Bardel J, Louwagie M, Jaquinod M, Jourdain A, Luche S, Rabilloud T, Macherel D, Garin J, Bourguignon J (2002) A survey of the plant mitochondrial proteome in relation to development. Proteomics 2: 880–898 - PubMed
-
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W, et al (1998) Analysis of 1.9 Mb contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 39: 809–821 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

