Dissecting the requirement for subgenomic promoter sequences by RNA recombination of brome mosaic virus in vivo: evidence for functional separation of transcription and recombination
- PMID: 15280464
- PMCID: PMC479100
- DOI: 10.1128/JVI.78.16.8552-8564.2004
Dissecting the requirement for subgenomic promoter sequences by RNA recombination of brome mosaic virus in vivo: evidence for functional separation of transcription and recombination
Abstract
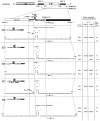
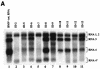
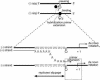
Previously, we and others mapped an increased homologous recombination activity within the subgenomic promoter (sgp) region in brome mosaic virus (BMV) RNA3. In order to correlate sgp-mediated recombination and transcription, in the present work we used BMV RNA3 constructs that carried altered sgp repeats. We observed that the removal or extension of the poly(U) tract reduced or increased recombination, respectively. Deletion of the sgp core hairpin or its replacement by a different stem-loop structure inhibited recombination activity. Nucleotide substitutions at the +1 or +2 transcription initiation position reduced recombination. The sgp core alone supported only basal recombination activity. The sites of crossovers mapped to the poly(U) region and to the core hairpin. The observed effects on recombination did not parallel those observed for transcription. To explain how both activities operate within the sgp sequence, we propose a dual mechanism whereby recombination is primed at the poly(U) tract by the predetached nascent plus strand, whereas transcription is initiated de novo at the sgp core.
Figures





Similar articles
-
A transcriptionally active subgenomic promoter supports homologous crossovers in a plus-strand RNA virus.J Virol. 2003 Jun;77(12):6769-76. doi: 10.1128/jvi.77.12.6769-6776.2003. J Virol. 2003. PMID: 12767997 Free PMC article.
-
Requirements for brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro.J Virol. 2004 Jun;78(12):6091-101. doi: 10.1128/JVI.78.12.6091-6101.2004. J Virol. 2004. PMID: 15163702 Free PMC article.
-
A conserved hairpin structure in Alfamovirus and Bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro.RNA. 2000 May;6(5):708-16. doi: 10.1017/s1355838200992471. RNA. 2000. PMID: 10836792 Free PMC article.
-
RNA recombination in brome mosaic virus, a model plus strand RNA virus.Acta Biochim Pol. 1998;45(4):847-68. Acta Biochim Pol. 1998. PMID: 10397334 Review.
-
The brome mosaic virus 3' untranslated sequence regulates RNA replication, recombination, and virion assembly.Virus Res. 2015 Aug 3;206:46-52. doi: 10.1016/j.virusres.2015.02.007. Epub 2015 Feb 14. Virus Res. 2015. PMID: 25687214 Review.
Cited by
-
Evidence for Internal Initiation of RNA Synthesis by the Hepatitis C Virus RNA-Dependent RNA Polymerase NS5B In Cellulo.J Virol. 2019 Sep 12;93(19):e00525-19. doi: 10.1128/JVI.00525-19. Print 2019 Oct 1. J Virol. 2019. PMID: 31315989 Free PMC article.
-
Mutations in the coat protein-binding cis-acting RNA motifs debilitate RNA recombination of Brome mosaic virus.Virus Res. 2012 Dec;170(1-2):138-49. doi: 10.1016/j.virusres.2012.10.001. Epub 2012 Oct 16. Virus Res. 2012. PMID: 23079110 Free PMC article.
-
Phylogenetic and Molecular Variability Studies Reveal a New Genetic Clade of Citrus leprosis virus C.Viruses. 2016 Jun 6;8(6):153. doi: 10.3390/v8060153. Viruses. 2016. PMID: 27275832 Free PMC article.
-
Co-infection with two strains of Brome mosaic bromovirus reveals common RNA recombination sites in different hosts.Virus Evol. 2015 Dec 23;1(1):vev021. doi: 10.1093/ve/vev021. eCollection 2015. Virus Evol. 2015. PMID: 27774290 Free PMC article.
-
Efficient in vitro system of homologous recombination in brome mosaic bromovirus.J Virol. 2006 Jun;80(12):6182-7. doi: 10.1128/JVI.02447-05. J Virol. 2006. PMID: 16731958 Free PMC article.
References
-
- Adkins, S., and C. C. Kao. 1998. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology 252:1-8. - PubMed
-
- Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. - PubMed
-
- Ahlquist, P., V. Luckow, and P. Kaesberg. 1981. Complete nucleotide sequence of brome mosaic virus RNA3. J. Mol. Biol. 153:23-38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

