Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors
- PMID: 15273297
- PMCID: PMC519198
- DOI: 10.1105/tpc.104.023309
Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors
Abstract
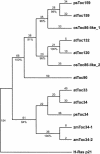
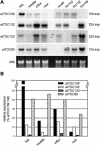
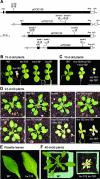
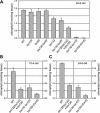
The initial stages of preprotein import into chloroplasts are mediated by the receptor GTPase Toc159. In Arabidopsis thaliana, Toc159 is encoded by a small gene family: atTOC159, atTOC132, atTOC120, and atTOC90. Phylogenetic analysis suggested that at least two distinct Toc159 subtypes, characterized by atToc159 and atToc132/atToc120, exist in plants. atTOC159 was strongly expressed in young, photosynthetic tissues, whereas atTOC132 and atTOC120 were expressed at a uniformly low level and so were relatively prominent in nonphotosynthetic tissues. Based on the albino phenotype of its knockout mutant, atToc159 was previously proposed to be a receptor with specificity for photosynthetic preproteins. To elucidate the roles of the other isoforms, we characterized Arabidopsis knockout mutants for each one. None of the single mutants had strong visible phenotypes, but toc132 toc120 double homozygotes appeared similar to toc159, indicating redundancy between atToc132 and atToc120. Transgenic complementation studies confirmed this redundancy but revealed little functional overlap between atToc132/atToc120 and atToc159 or atToc90. Unlike toc159, toc132 toc120 caused structural abnormalities in root plastids. Furthermore, when proteomics and transcriptomics were used to compare toc132 with ppi1 (a receptor mutant that is specifically defective in the expression, import, and accumulation of photosynthetic proteins), major differences were observed, suggesting that atToc132 (and atToc120) has specificity for nonphotosynthetic proteins. When both atToc159 and the major isoform of the other subtype, atToc132, were absent, an embryo-lethal phenotype resulted, demonstrating the essential role of Toc159 in the import mechanism.
Figures


Similar articles
-
A split-ubiquitin yeast two-hybrid screen to examine the substrate specificity of atToc159 and atToc132, two Arabidopsis chloroplast preprotein import receptors.PLoS One. 2014 Apr 15;9(4):e95026. doi: 10.1371/journal.pone.0095026. eCollection 2014. PLoS One. 2014. PMID: 24736607 Free PMC article.
-
Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids.Mol Biol Cell. 2004 Jul;15(7):3379-92. doi: 10.1091/mbc.e03-12-0923. Epub 2004 Apr 16. Mol Biol Cell. 2004. PMID: 15090618 Free PMC article.
-
The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts.Plant Cell. 2010 Jun;22(6):1947-60. doi: 10.1105/tpc.110.074328. Epub 2010 Jun 18. Plant Cell. 2010. PMID: 20562235 Free PMC article.
-
The TOC GTPase Receptors: Regulators of the Fidelity, Specificity and Substrate Profiles of the General Protein Import Machinery of Chloroplasts.Protein J. 2019 Jun;38(3):343-350. doi: 10.1007/s10930-019-09846-3. Protein J. 2019. PMID: 31201619 Free PMC article. Review.
-
Protein transport in organelles: The Toc complex way of preprotein import.FEBS J. 2009 Mar;276(5):1156-65. doi: 10.1111/j.1742-4658.2009.06873.x. FEBS J. 2009. PMID: 19187236 Review.
Cited by
-
Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts.Plant Cell. 2010 May;22(5):1516-31. doi: 10.1105/tpc.109.071415. Epub 2010 May 18. Plant Cell. 2010. PMID: 20484004 Free PMC article.
-
A new member of the psToc159 family contributes to distinct protein targeting pathways in pea chloroplasts.Front Plant Sci. 2014 May 28;5:239. doi: 10.3389/fpls.2014.00239. eCollection 2014. Front Plant Sci. 2014. PMID: 24904628 Free PMC article.
-
Evolution of rubisco complex small subunit transit peptides from algae to plants.Sci Rep. 2017 Aug 24;7(1):9279. doi: 10.1038/s41598-017-09473-x. Sci Rep. 2017. PMID: 28839179 Free PMC article.
-
Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins.Plant Cell. 2011 Nov;23(11):3911-28. doi: 10.1105/tpc.111.092882. Epub 2011 Nov 29. Plant Cell. 2011. PMID: 22128122 Free PMC article.
-
ARSENATE INDUCED CHLOROSIS 1/ TRANSLOCON AT THE OUTER ENVOLOPE MEMBRANE OF CHLOROPLASTS 132 Protects Chloroplasts from Arsenic Toxicity.Plant Physiol. 2018 Dec;178(4):1568-1583. doi: 10.1104/pp.18.01042. Epub 2018 Oct 11. Plant Physiol. 2018. PMID: 30309965 Free PMC article.
References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
-
- Aronsson, H., Combe, J., and Jarvis, P. (2003). Unusual nucleotide-binding properties of the chloroplast protein import receptor, atToc33. FEBS Lett. 544, 79–85. - PubMed
-
- Aronsson, H., and Jarvis, P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529, 215–220. - PubMed
-
- Baker, K.P., and Schatz, G. (1991). Mitochondrial proteins essential for viability mediate protein import into yeast mitochondria. Nature 349, 205–208. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

