Analysis of splice variants of the immediate-early 1 region of human cytomegalovirus
- PMID: 15254190
- PMCID: PMC446109
- DOI: 10.1128/JVI.78.15.8191-8200.2004
Analysis of splice variants of the immediate-early 1 region of human cytomegalovirus
Abstract
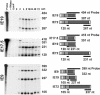
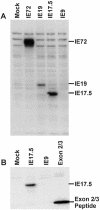
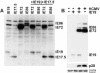
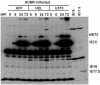
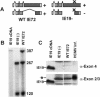
The major immediate-early (MIE) gene of human cytomegalovirus (HCMV) produces multiple mRNAs through differential splicing and polyadenylation. Reverse transcriptase PCR was used to characterize transcripts from exons 1, 2, 3, and 4 (immediate-early 1 [IE1]). The expected IE72 and IE19 mRNAs were detected, as well as two heretofore-uncharacterized transcripts designated IE17.5 and IE9. The IE72, IE19, and IE17.5 transcripts utilized the same 5'-splice site in exon 3. IE9 utilized a cryptic 5'-splice site within exon 3. The IE19, IE17.5, and IE9 transcripts all used different 3'-splice sites within exon 4. These spliced species occur in infected human foreskin fibroblast (HFF) cells, with accumulation kinetics similar to those of IE72 mRNA. IE19 and IE9 RNAs were much more abundant than IE17.5 RNA. Transfection of CV-1 cells with cDNAs resulted in IE19 and IE17.5 proteins detectable by antibodies to either N-terminal or C-terminal epitopes. No IE9 protein product has been detected. We have not been able to detect IE19, IE17.5, or IE9 proteins during infection of HFF, HEL, or U373MG cells. Failure to detect IE19 protein contrasts with a previous report (M. Shirakata, M. Terauchi, M. Ablikin, K. Imadome, K. Hirai, T. Aso, and Y. Yamanashi, J. Virol. 76:3158-3167, 2002) of IE19 protein expression in HCMV-infected HEL cells. Our analysis suggests that an N-terminal breakdown product of IE72 may be mistaken for IE19. Expression of IE19 or IE17.5 from its respective cDNA results in repression of viral gene expression in infected cells. We speculate that expression of these proteins during infection may be restricted to specific conditions or cell types.
Figures




Similar articles
-
Novel immediate-early protein IE19 of human cytomegalovirus activates the origin recognition complex I promoter in a cooperative manner with IE72.J Virol. 2002 Apr;76(7):3158-67. doi: 10.1128/jvi.76.7.3158-3167.2002. J Virol. 2002. PMID: 11884540 Free PMC article.
-
Human cytomegalovirus late protein encoded by ie2: a trans-activator as well as a repressor of gene expression.J Gen Virol. 1994 Sep;75 ( Pt 9):2337-48. doi: 10.1099/0022-1317-75-9-2337. J Gen Virol. 1994. PMID: 8077932
-
Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins.J Virol. 1990 Apr;64(4):1498-506. doi: 10.1128/JVI.64.4.1498-1506.1990. J Virol. 1990. PMID: 2157038 Free PMC article.
-
Human cytomegalovirus UL37 immediate early target minigene RNAs are accurately spliced and polyadenylated.J Gen Virol. 2003 Jan;84(Pt 1):29-39. doi: 10.1099/vir.0.18700-0. J Gen Virol. 2003. PMID: 12533698
-
Transcription factor Sp1 mediates cell-specific trans-activation of the human cytomegalovirus DNA polymerase gene promoter by immediate-early protein IE86 in glioblastoma U373MG cells.J Virol. 1998 Jan;72(1):236-44. doi: 10.1128/JVI.72.1.236-244.1998. J Virol. 1998. PMID: 9420220 Free PMC article.
Cited by
-
Two Polypyrimidine Tracts in Intron 4 of the Major Immediate Early Gene Are Critical for Gene Expression Switching from IE1 to IE2 and for Replication of Human Cytomegalovirus.J Virol. 2016 Jul 27;90(16):7339-7349. doi: 10.1128/JVI.00837-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27252533 Free PMC article.
-
The human cytomegalovirus decathlon: Ten critical replication events provide opportunities for restriction.Front Cell Dev Biol. 2022 Nov 25;10:1053139. doi: 10.3389/fcell.2022.1053139. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506089 Free PMC article. Review.
-
Antisense transcription in the human cytomegalovirus transcriptome.J Virol. 2007 Oct;81(20):11267-81. doi: 10.1128/JVI.00007-07. Epub 2007 Aug 8. J Virol. 2007. PMID: 17686857 Free PMC article.
-
The IE2 60-kilodalton and 40-kilodalton proteins are dispensable for human cytomegalovirus replication but are required for efficient delayed early and late gene expression and production of infectious virus.J Virol. 2007 Mar;81(6):2573-83. doi: 10.1128/JVI.02454-06. Epub 2007 Jan 3. J Virol. 2007. PMID: 17202222 Free PMC article.
-
High-resolution human cytomegalovirus transcriptome.Proc Natl Acad Sci U S A. 2011 Dec 6;108(49):19755-60. doi: 10.1073/pnas.1115861108. Epub 2011 Nov 22. Proc Natl Acad Sci U S A. 2011. PMID: 22109557 Free PMC article.
References
-
- Baracchini, E., E. Glezer, K. Fish, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1992. An isoform variant of the cytomegalovirus immediate-early auto repressor functions as a transcriptional activator. Virology 188:518-529. - PubMed
-
- Ghazal, P., J. Young, E. Giulietti, C. DeMattei, J. Garcia, R. Gaynor, R. M. Stenberg, and J. A. Nelson. 1991. A discrete cis element in the human immunodeficiency virus long terminal repeat mediates synergistic trans activation by cytomegalovirus immediate-early proteins. J. Virol. 65:6735-6742. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

