Regulation of relative abundance of arterivirus subgenomic mRNAs
- PMID: 15254182
- PMCID: PMC446141
- DOI: 10.1128/JVI.78.15.8102-8113.2004
Regulation of relative abundance of arterivirus subgenomic mRNAs
Abstract
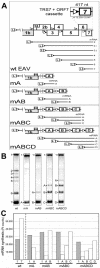
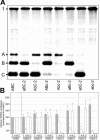
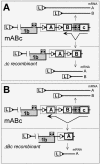
The subgenomic (sg) mRNAs of arteriviruses (order Nidovirales) form a 5'- and 3'-coterminal nested set with the viral genome. Their 5' common leader sequence is derived from the genomic 5'-proximal region. Fusion of sg RNA leader and "body" segments involves a discontinuous transcription step. Presumably during minus-strand synthesis, the nascent RNA strand is transferred from one site in the genomic template to another, a process guided by conserved transcription-regulating sequences (TRSs) at these template sites. Subgenomic RNA species are produced in different but constant molar ratios, with the smallest RNAs usually being most abundant. Factors thought to influence sg RNA synthesis are size differences between sg RNA species, differences in sequence context between body TRSs, and the mutual influence (or competition) between strand transfer reactions occurring at different body TRSs. Using an Equine arteritis virus infectious cDNA clone, we investigated how body TRS activity affected sg RNA synthesis from neighboring body TRSs. Flanking sequences were standardized by head-to-tail insertion of several copies of an RNA7 body TRS cassette. A perfect gradient of sg RNA abundance, progressively favoring smaller RNA species, was observed. Disruption of body TRS function by mutagenesis did not have a significant effect on the activity of other TRSs. However, deletion of body TRS-containing regions enhanced synthesis of sg RNAs from upstream TRSs but not of those produced from downstream TRSs. The results of this study provide considerable support for the proposed discontinuous extension of minus-strand RNA synthesis as a crucial step in sg RNA synthesis.
Figures
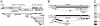


Similar articles
-
Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region.J Virol. 2005 May;79(10):6312-24. doi: 10.1128/JVI.79.10.6312-6324.2005. J Virol. 2005. PMID: 15858015 Free PMC article.
-
The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis.J Virol. 2003 Jan;77(2):1175-83. doi: 10.1128/jvi.77.2.1175-1183.2003. J Virol. 2003. PMID: 12502834 Free PMC article.
-
Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis.EMBO J. 2001 Dec 17;20(24):7220-8. doi: 10.1093/emboj/20.24.7220. EMBO J. 2001. PMID: 11742998 Free PMC article.
-
Subgenomic messenger RNAs: mastering regulation of (+)-strand RNA virus life cycle.Virology. 2011 Apr 10;412(2):245-55. doi: 10.1016/j.virol.2011.02.007. Epub 2011 Mar 5. Virology. 2011. PMID: 21377709 Free PMC article. Review.
-
Nidovirus transcription: how to make sense...?J Gen Virol. 2006 Jun;87(Pt 6):1403-1421. doi: 10.1099/vir.0.81611-0. J Gen Virol. 2006. PMID: 16690906 Review.
Cited by
-
PRRSV structure, replication and recombination: Origin of phenotype and genotype diversity.Virology. 2015 May;479-480:475-86. doi: 10.1016/j.virol.2015.02.012. Epub 2015 Mar 7. Virology. 2015. PMID: 25759097 Free PMC article. Review.
-
Glycogen synthase kinase-3: A putative target to combat severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic.Cytokine Growth Factor Rev. 2021 Apr;58:92-101. doi: 10.1016/j.cytogfr.2020.08.002. Epub 2020 Aug 25. Cytokine Growth Factor Rev. 2021. PMID: 32948440 Free PMC article. Review.
-
Reverse genetic manipulation of the overlapping coding regions for structural proteins of the type II porcine reproductive and respiratory syndrome virus.Virology. 2009 Jan 5;383(1):22-31. doi: 10.1016/j.virol.2008.09.013. Epub 2008 Nov 5. Virology. 2009. PMID: 18977502 Free PMC article.
-
Gene Variations in Cis-Acting Elements between the Taiwan and Prototype Strains of Porcine Epidemic Diarrhea Virus Alter Viral Gene Expression.Genes (Basel). 2018 Nov 29;9(12):591. doi: 10.3390/genes9120591. Genes (Basel). 2018. PMID: 30501108 Free PMC article.
-
Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis.J Virol. 2005 Feb;79(4):2506-16. doi: 10.1128/JVI.79.4.2506-2516.2005. J Virol. 2005. PMID: 15681451 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

