Roles of Bim in apoptosis of normal and Bcr-Abl-expressing hematopoietic progenitors
- PMID: 15226421
- PMCID: PMC434248
- DOI: 10.1128/MCB.24.14.6172-6183.2004
Roles of Bim in apoptosis of normal and Bcr-Abl-expressing hematopoietic progenitors
Abstract
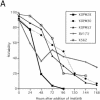
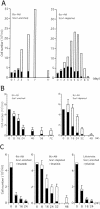
Bcr-Abl kinase is known to reverse apoptosis of cytokine-dependent cells due to cytokine deprivation, although it has been controversial whether chronic myeloid leukemia (CML) progenitors have the potential to survive under conditions in which there are limited amounts of cytokines. Here we demonstrate that early hematopoietic progenitors (Sca-1(+) c-Kit(+) Lin(-)) isolated from normal mice rapidly undergo apoptosis in the absence of cytokines. In these cells, the expression of Bim, a proapoptotic relative of Bcl-2 which plays a key role in the cytokine-mediated survival system, is induced. In contrast, those cells isolated from our previously established CML model mice resist apoptosis in cytokine-free medium without the induction of Bim expression, and these effects are reversed by the Abl-specific kinase inhibitor imatinib mesylate. In addition, the expression levels of Bim are uniformly low in cell lines established from patients in the blast crisis phase of CML, and imatinib induced Bim in these cells. Moreover, small interfering RNA that reduces the expression level of Bim effectively rescues CML cells from apoptosis caused by imatinib. These findings suggest that Bim plays an important role in the apoptosis of early hematopoietic progenitors and that Bcr-Abl supports cell survival in part through downregulation of this cell death activator.
Figures




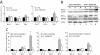
Similar articles
-
Low-level expression of proapoptotic Bcl-2-interacting mediator in leukemic cells in patients with chronic myeloid leukemia: role of BCR/ABL, characterization of underlying signaling pathways, and reexpression by novel pharmacologic compounds.Cancer Res. 2005 Oct 15;65(20):9436-44. doi: 10.1158/0008-5472.CAN-05-0972. Cancer Res. 2005. PMID: 16230407
-
Chronic myeloid leukaemia: an investigation into the role of Bcr-Abl-induced abnormalities in glucose transport regulation.Oncogene. 2005 May 5;24(20):3257-67. doi: 10.1038/sj.onc.1208461. Oncogene. 2005. PMID: 15735728
-
Role of SPA-1 in phenotypes of chronic myelogenous leukemia induced by BCR-ABL-expressing hematopoietic progenitors in a mouse model.Cancer Res. 2006 Oct 15;66(20):9967-76. doi: 10.1158/0008-5472.CAN-06-1346. Cancer Res. 2006. PMID: 17047059
-
[Chemokines in chronic myeloid leukemia].Rinsho Ketsueki. 2016 Feb;57(2):129-36. doi: 10.11406/rinketsu.57.129. Rinsho Ketsueki. 2016. PMID: 26935630 Review. Japanese.
-
Cytokine-mediated cell survival.Int J Hematol. 2004 Oct;80(3):210-4. doi: 10.1532/ijh97.04093. Int J Hematol. 2004. PMID: 15540894 Review.
Cited by
-
BH3 mimetics in combination with nilotinib or ponatinib represent a promising therapeutic strategy in blast phase chronic myeloid leukemia.Cell Death Discov. 2022 Nov 15;8(1):457. doi: 10.1038/s41420-022-01211-1. Cell Death Discov. 2022. PMID: 36379918 Free PMC article.
-
Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review.Transl Lung Cancer Res. 2015 Feb;4(1):67-81. doi: 10.3978/j.issn.2218-6751.2014.11.06. Transl Lung Cancer Res. 2015. PMID: 25806347 Free PMC article. Review.
-
The protein kinase A pathway-regulated transcriptome of endometrial stromal fibroblasts reveals compromised differentiation and persistent proliferative potential in endometriosis.Endocrinology. 2010 Mar;151(3):1341-55. doi: 10.1210/en.2009-0923. Epub 2010 Jan 12. Endocrinology. 2010. PMID: 20068008 Free PMC article.
-
The application of BH3 mimetics in myeloid leukemias.Cell Death Dis. 2021 Feb 26;12(2):222. doi: 10.1038/s41419-021-03500-6. Cell Death Dis. 2021. PMID: 33637708 Free PMC article. Review.
-
Requirement for antiapoptotic MCL-1 in the survival of BCR-ABL B-lineage acute lymphoblastic leukemia.Blood. 2013 Aug 29;122(9):1587-98. doi: 10.1182/blood-2012-06-440230. Epub 2013 Jul 23. Blood. 2013. PMID: 23881917 Free PMC article.
References
-
- Adams, J. M., and S. Cory. 1998. The Bcl-2 protein family: arbiters of cell survival. Science 281:1322-1326. - PubMed
-
- Akiyama, T., P. Bouillet, T. Miyazaki, Y. Kadono, H. Chikuda, U. I. Chung, A. Fukuda, A. Hikita, H. Seto, T. Okada, T. Inaba, A. Sanjay, R. Baron, H. Kawaguchi, H. Oda, K. Nakamura, A. Strasser, and S. Tanaka. 2003. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J. 22:6653-6664. - PMC - PubMed
-
- Amos, T. A., J. L. Lewis, F. H. Grand, R. P. Gooding, J. M. Goldman, and M. Y. Gordon. 1995. Apoptosis in chronic myeloid leukaemia: normal responses by progenitor cells to growth factor deprivation, X-irradiation and glucocorticoids. Br. J. Haematol. 91:387-393. - PubMed
-
- Bedi, A., B. A. Zehnbauer, J. P. Barber, S. J. Sharkis, and R. J. Jones. 1994. Inhibition of apoptosis by BCR-ABL in chronic myeloid leukemia. Blood 83:2038-2044. - PubMed
-
- Biswas, S. C., and L. A. Greene. 2002. Nerve growth factor (NGF) down-regulates the Bcl-2 homology 3 (BH3) domain-only protein Bim and suppresses its proapoptotic activity by phosphorylation. J. Biol. Chem. 277:49511-49516. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
