Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes
- PMID: 15192704
- PMCID: PMC449771
- DOI: 10.1038/sj.emboj.7600261
Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes
Abstract
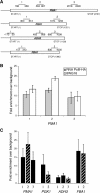
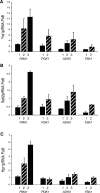
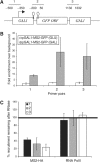
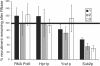
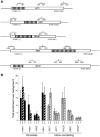
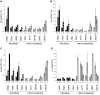
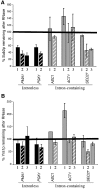
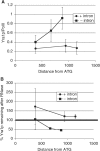
The TREX complex is involved in both transcription elongation and mRNA export and is recruited to nascent transcription complexes. We have examined Yra1p, Sub2p and Hpr1p recruitment to nine genes of varying lengths and transcription frequencies. All three proteins increase from the 5' to the 3' ends of the four intronless genes examined. A modified chromatin immunoprecipitation assay that includes an RNase step indicates that Sub2p is bound to nascent RNA, Yra1p is associated with both RNA and DNA, and Hpr1p is associated with DNA. Although Hpr1p is recruited similarly to both intronless and intron-containing genes, low Yra1p and Sub2p levels are present on a subset of intron-containing genes. The residual Yra1p and Sub2p recruitment is less RNA-associated, and this correlates with high levels of U1 SnRNP on these genes. These experiments support a model in which TREX is recruited via the transcription machinery and then Yra1p and Sub2p are transferred to the nascent RNA. On some intron-containing genes, retention and/or transfer of Yra1p and Sub2p to nascent RNA are inhibited.
Figures
Similar articles
-
Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p.Mol Cell Biol. 2002 Dec;22(23):8241-53. doi: 10.1128/MCB.22.23.8241-8253.2002. Mol Cell Biol. 2002. PMID: 12417727 Free PMC article.
-
Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism.RNA. 2002 Aug;8(8):969-80. doi: 10.1017/s1355838202020046. RNA. 2002. PMID: 12212852 Free PMC article.
-
Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p.Nature. 2001 Oct 11;413(6856):648-52. doi: 10.1038/35098113. Nature. 2001. PMID: 11675790
-
An intron in the YRA1 gene is required to control Yra1 protein expression and mRNA export in yeast.EMBO Rep. 2002 May;3(5):438-42. doi: 10.1093/embo-reports/kvf091. Epub 2002 Apr 18. EMBO Rep. 2002. PMID: 11964382 Free PMC article.
-
Nuclear export of messenger RNA.Results Probl Cell Differ. 2002;35:133-50. doi: 10.1007/978-3-540-44603-3_7. Results Probl Cell Differ. 2002. PMID: 11791404 Review. No abstract available.
Cited by
-
Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor.EMBO J. 2012 Mar 21;31(6):1605-16. doi: 10.1038/emboj.2012.10. Epub 2012 Feb 7. EMBO J. 2012. PMID: 22314234 Free PMC article.
-
Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud.Genes Dev. 2010 Sep 1;24(17):1914-26. doi: 10.1101/gad.1937510. Epub 2010 Aug 16. Genes Dev. 2010. PMID: 20713510 Free PMC article.
-
Mechanisms of nuclear mRNA export: A structural perspective.Traffic. 2019 Nov;20(11):829-840. doi: 10.1111/tra.12691. Epub 2019 Sep 12. Traffic. 2019. PMID: 31513326 Free PMC article. Review.
-
Transcriptional factor ENY2 promotes recruitment of the THO complex to the hsp70 gene of Drosophila melanogaster.Dokl Biochem Biophys. 2010 Sep-Oct;434:227-31. doi: 10.1134/S1607672910050029. Epub 2010 Oct 20. Dokl Biochem Biophys. 2010. PMID: 20960243 No abstract available.
-
Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes.Mol Cell Biol. 2006 Nov;26(21):7858-70. doi: 10.1128/MCB.00870-06. Epub 2006 Sep 5. Mol Cell Biol. 2006. PMID: 16954382 Free PMC article.
References
-
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 - PubMed
-
- Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834 - PMC - PubMed
-
- Dye MJ, Proudfoot NJ (1999) Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell 3: 371–378 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

