A molecular dynamics study of reovirus attachment protein sigma1 reveals conformational changes in sigma1 structure
- PMID: 15189844
- PMCID: PMC1304249
- DOI: 10.1529/biophysj.103.030825
A molecular dynamics study of reovirus attachment protein sigma1 reveals conformational changes in sigma1 structure
Abstract
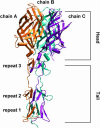
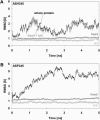
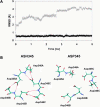
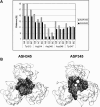
Molecular dynamics simulations were performed using the recently determined crystal structure of the reovirus attachment protein, sigma1. These studies were conducted to improve an understanding of two unique features of sigma1 structure: the protonation state of Asp(345), which is buried in the sigma1 trimer interface, and the flexibility of the protein at a defined region below the receptor-binding head domain. Three copies of aspartic acids Asp(345) and Asp(346) cluster in a solvent-inaccessible and hydrophobic region at the sigma1 trimer interface. These residues are hypothesized to mediate conformational changes in sigma1 during viral attachment or cell entry. Our results indicate that protonation of Asp(345) is essential to the integrity of the trimeric structure seen by x-ray crystallography, whereas deprotonation induces structural changes that destabilize the trimer interface. This finding was confirmed by electrostatic calculations using the finite difference Poisson-Boltzmann method. Earlier studies show that sigma1 can exist in retracted and extended conformations on the viral surface. Since protonated Asp(345) is necessary to form a stable, extended trimer, our results suggest that protonation of Asp(345) may allow for a structural transition from a partially detrimerized molecule to the fully formed trimer seen in the crystal structure. Additional studies were conducted to quantify the previously observed flexibility of sigma1 at a defined region below the receptor-binding head domain. Increased mobility was observed for three polar residues (Ser(291), Thr(292), and Ser(293)) located within an insertion between the second and third beta-spiral repeats of the crystallized portion of the sigma1 tail. These amino acids interact with water molecules of the solvent bulk and are responsible for oscillating movement of the head of approximately 50 degrees during 5 ns of simulations. This flexibility may facilitate viral attachment and also function in cell entry and disassembly. These findings provide new insights about the conformational dynamics of sigma1 that likely underlie the initiation of the reovirus infectious cycle.
Figures

Similar articles
-
An Unusual Aspartic Acid Cluster in the Reovirus Attachment Fiber σ1 Mediates Stability at Low pH and Preserves Trimeric Organization.J Virol. 2022 Apr 27;96(8):e0033122. doi: 10.1128/jvi.00331-22. Epub 2022 Apr 5. J Virol. 2022. PMID: 35380459 Free PMC article.
-
Structural and Functional Features of the Reovirus σ1 Tail.J Virol. 2018 Jun 29;92(14):e00336-18. doi: 10.1128/JVI.00336-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29695426 Free PMC article.
-
Crystal structure of reovirus attachment protein sigma1 reveals evolutionary relationship to adenovirus fiber.EMBO J. 2002 Jan 15;21(1-2):1-11. doi: 10.1093/emboj/21.1.1. EMBO J. 2002. PMID: 11782420 Free PMC article.
-
Structural similarities in the cellular receptors used by adenovirus and reovirus.Viral Immunol. 2004;17(2):129-43. doi: 10.1089/0882824041310621. Viral Immunol. 2004. PMID: 15279694 Review.
-
Avian reovirus: structure and biology.Virus Res. 2007 Feb;123(2):105-19. doi: 10.1016/j.virusres.2006.09.005. Epub 2006 Oct 2. Virus Res. 2007. PMID: 17018239 Review.
Cited by
-
From touchdown to transcription: the reovirus cell entry pathway.Curr Top Microbiol Immunol. 2010;343:91-119. doi: 10.1007/82_2010_32. Curr Top Microbiol Immunol. 2010. PMID: 20397070 Free PMC article. Review.
-
An Unusual Aspartic Acid Cluster in the Reovirus Attachment Fiber σ1 Mediates Stability at Low pH and Preserves Trimeric Organization.J Virol. 2022 Apr 27;96(8):e0033122. doi: 10.1128/jvi.00331-22. Epub 2022 Apr 5. J Virol. 2022. PMID: 35380459 Free PMC article.
-
Structural and Functional Features of the Reovirus σ1 Tail.J Virol. 2018 Jun 29;92(14):e00336-18. doi: 10.1128/JVI.00336-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29695426 Free PMC article.
References
-
- Barton, E. S., J. C. Forrest, J. L. Connolly, J. D. Chappell, Y. Liu, F. J. Schnell, A. Nusrat, C. A. Parkos, and T. S. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell. 104:441–451. - PubMed
-
- Berendsen, H. J. C., J. P. M. Postma, W. F. Van Gunsteren, A. Di Nola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690.
-
- Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994a. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 371:37–43. - PubMed
-
- Bullough, P. A., F. M. Hughson, A. C. Treharne, R. W. Ruigrok, J. J. Skehel, and D. C. Wiley. 1994b. Crystals of a fragment of influenza haemagglutinin in the low pH induced conformation. J. Mol. Biol. 236:1262–1265. - PubMed
-
- Case, D. A., D. A. Pearlman, J. W. Caldwell, T. E. Cheatham III, W. S. Ross, C. L. Simmerling, T. A. Darden, K. M. Merz, R. V. Stanton, J. J. Vincent, M. Crowley, D. M. Ferguson, R. J. Radmer, G. L. Seibel, U. C. Singh, P. K. Weiner, and P. A. Kollman. 1999. AMBER 6. University of California, San Francisco, CA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

