"Sleeping beauty": quiescence in Saccharomyces cerevisiae
- PMID: 15187181
- PMCID: PMC419917
- DOI: 10.1128/MMBR.68.2.187-206.2004
"Sleeping beauty": quiescence in Saccharomyces cerevisiae
Abstract
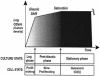
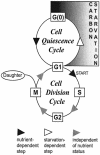
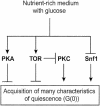
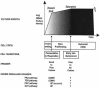
The cells of organisms as diverse as bacteria and humans can enter stable, nonproliferating quiescent states. Quiescent cells of eukaryotic and prokaryotic microorganisms can survive for long periods without nutrients. This alternative state of cells is still poorly understood, yet much benefit is to be gained by understanding it both scientifically and with reference to human health. Here, we review our knowledge of one "model" quiescent cell population, in cultures of yeast grown to stationary phase in rich media. We outline the importance of understanding quiescence, summarize the properties of quiescent yeast cells, and clarify some definitions of the state. We propose that the processes by which a cell enters into, maintains viability in, and exits from quiescence are best viewed as an environmentally triggered cycle: the cell quiescence cycle. We synthesize what is known about the mechanisms by which yeast cells enter into quiescence, including the possible roles of the protein kinase A, TOR, protein kinase C, and Snf1p pathways. We also discuss selected mechanisms by which quiescent cells maintain viability, including metabolism, protein modification, and redox homeostasis. Finally, we outline what is known about the process by which cells exit from quiescence when nutrients again become available.
Figures
Similar articles
-
Stationary phase in yeast.Curr Opin Microbiol. 2002 Dec;5(6):602-7. doi: 10.1016/s1369-5274(02)00377-6. Curr Opin Microbiol. 2002. PMID: 12457705 Review.
-
Transcriptional regulation in yeast during diauxic shift and stationary phase.OMICS. 2010 Dec;14(6):629-38. doi: 10.1089/omi.2010.0069. Epub 2010 Sep 23. OMICS. 2010. PMID: 20863251 Free PMC article. Review.
-
Genetic interaction profiles of regulatory kinases differ between environmental conditions and cellular states.Mol Syst Biol. 2020 May;16(5):e9167. doi: 10.15252/msb.20199167. Mol Syst Biol. 2020. PMID: 32449603 Free PMC article.
-
Preparation and Analysis of Saccharomyces cerevisiae Quiescent Cells.Methods Mol Biol. 2018;1686:125-135. doi: 10.1007/978-1-4939-7371-2_9. Methods Mol Biol. 2018. PMID: 29030817
-
Cellular quiescence in budding yeast.Yeast. 2021 Jan;38(1):12-29. doi: 10.1002/yea.3545. Epub 2021 Jan 25. Yeast. 2021. PMID: 33350503 Free PMC article. Review.
Cited by
-
The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan.PLoS Genet. 2015 Jun 23;11(6):e1005282. doi: 10.1371/journal.pgen.1005282. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26103122 Free PMC article.
-
Hematopoietic stem cell stretches and moves in its bone marrow niche.Crit Rev Oncol Hematol. 2021 Jul;163:103368. doi: 10.1016/j.critrevonc.2021.103368. Epub 2021 May 26. Crit Rev Oncol Hematol. 2021. PMID: 34051302 Free PMC article. Review.
-
Eukaryotic Adaptation to Years-Long Starvation Resembles that of Bacteria.iScience. 2019 Sep 27;19:545-558. doi: 10.1016/j.isci.2019.08.002. Epub 2019 Aug 8. iScience. 2019. PMID: 31470363 Free PMC article.
-
Cytocidal amino acid starvation of Saccharomyces cerevisiae and Candida albicans acetolactate synthase (ilv2{Delta}) mutants is influenced by the carbon source and rapamycin.Microbiology (Reading). 2010 Mar;156(Pt 3):929-939. doi: 10.1099/mic.0.034348-0. Epub 2009 Dec 17. Microbiology (Reading). 2010. PMID: 20019084 Free PMC article.
-
Sterol Metabolism Differentially Contributes to Maintenance and Exit of Quiescence.Front Cell Dev Biol. 2022 Feb 14;10:788472. doi: 10.3389/fcell.2022.788472. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35237594 Free PMC article.
References
-
- Ashrafi, K., T. A. Farazi, and J. I. Gordon. 1998. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J. Biol. Chem. 273:25864-25874. - PubMed
-
- Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. - PubMed
-
- Benaroudj, N., D. H. Lee, and A. L. Goldberg. 2001. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 276:24261-24267. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

