Purification and characterization of enzymes exhibiting beta-D-xylosidase activities in stem tissues of Arabidopsis
- PMID: 15181203
- PMCID: PMC514122
- DOI: 10.1104/pp.104.041269
Purification and characterization of enzymes exhibiting beta-D-xylosidase activities in stem tissues of Arabidopsis
Abstract
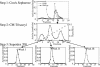
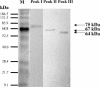
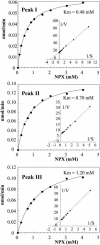
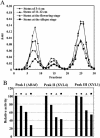
This work describes the purification and characterization of enzymes that exhibit beta-d-xylosidase activity in stem tissues of Arabidopsis. This is the first detailed investigation that concerns the characterization of catalytic properties and sequence identity of enzymes with beta-D-xylosidase activities in a dicotyledonous plant. Three different enzymes, ARAf, XYL4, and XYL1 with apparent molecular masses of 75, 67, and 64 kD, respectively, were purified to homogeneity. ARAf was identified as a putative alpha-L-arabinofuranosidase, and XYL4 and XYL1 as putative beta-D-xylosidases using matrix-assisted laser-desorption ionization time of flight. ARAf belongs to family 51 and XYL4 and XYL1 to family 3 of glycoside hydrolases. ARAf and XYL1 have highest specificity for p-nitrophenyl-alpha-L-arabinofuranoside and XYL4 for p-nitrophenyl-beta-D-xylopyranoside and natural substrates such as xylobiose and xylotetraose. XYL4 was shown to release mainly D-Xyl from oat spelt xylan, rye arabinoxylan, wheat arabinoxylan, and oligoarabinoxylans. ARAf and XYL1 can also release D-Xyl from these substrates but less efficiently than XYL4. Moreover, they can also release L-Ara from arabinoxylans and arabinan. Overall, the results indicate that XYL4 possesses enzymatic specificity characteristic for a beta-D-xylosidase, while ARAf and XYL1 act as bifunctional alpha-L-arabinofuranosidase/beta-D-xylosidases. Analysis of the activity of these three enzymes in stem tissues at different stages of development has shown that young stems possess the highest activities for all three enzymes in comparison to the activities of the enzymes present in stems at older stages of development. High enzyme activities are most likely related to the necessary modifications of cell wall structure occurring during plant growth.
Figures


Similar articles
-
Purification, functional characterization, cloning, and identification of mutants of a seed-specific arabinan hydrolase in Arabidopsis.J Exp Bot. 2006;57(10):2339-51. doi: 10.1093/jxb/erj205. Epub 2006 Jun 23. J Exp Bot. 2006. PMID: 16798843
-
Cloning of genes encoding alpha-L-arabinofuranosidase and beta-xylosidase from Trichoderma reesei by expression in Saccharomyces cerevisiae.Appl Environ Microbiol. 1996 Oct;62(10):3840-6. doi: 10.1128/aem.62.10.3840-3846.1996. Appl Environ Microbiol. 1996. PMID: 8837440 Free PMC article.
-
The construction and characterization of two xylan-degrading chimeric enzymes.Biotechnol Bioeng. 2009 Feb 15;102(3):684-92. doi: 10.1002/bit.22112. Biotechnol Bioeng. 2009. PMID: 18828173
-
Penicillium purpurogenum produces two GH family 43 enzymes with β-xylosidase activity, one monofunctional and the other bifunctional: Biochemical and structural analyses explain the difference.Arch Biochem Biophys. 2013 Dec;540(1-2):117-24. doi: 10.1016/j.abb.2013.10.017. Epub 2013 Oct 31. Arch Biochem Biophys. 2013. PMID: 24184421
-
Structure and function analysis of Pseudomonas plant cell wall hydrolases.Biochem Soc Trans. 1998 May;26(2):185-90. doi: 10.1042/bst0260185. Biochem Soc Trans. 1998. PMID: 9649745 Review. No abstract available.
Cited by
-
Biochemical analysis of a beta-D-xylosidase and a bifunctional xylanase-ferulic acid esterase from a xylanolytic gene cluster in Prevotella ruminicola 23.J Bacteriol. 2009 May;191(10):3328-38. doi: 10.1128/JB.01628-08. Epub 2009 Mar 20. J Bacteriol. 2009. PMID: 19304844 Free PMC article.
-
Forgotten Actors: Glycoside Hydrolases During Elongation Growth of Maize Primary Root.Front Plant Sci. 2022 Feb 10;12:802424. doi: 10.3389/fpls.2021.802424. eCollection 2021. Front Plant Sci. 2022. PMID: 35222452 Free PMC article.
-
Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought.BMC Genomics. 2007 Sep 2;8:303. doi: 10.1186/1471-2164-8-303. BMC Genomics. 2007. PMID: 17764573 Free PMC article.
-
Cell wall modifications in Arabidopsis plants with altered alpha-L-arabinofuranosidase activity.Plant Physiol. 2008 May;147(1):63-77. doi: 10.1104/pp.107.110023. Epub 2008 Mar 14. Plant Physiol. 2008. PMID: 18344421 Free PMC article.
-
Increase in cellulose accumulation and improvement of saccharification by overexpression of arabinofuranosidase in rice.PLoS One. 2013 Nov 4;8(11):e78269. doi: 10.1371/journal.pone.0078269. eCollection 2013. PLoS One. 2013. PMID: 24223786 Free PMC article.
References
-
- Atkins GL, Nimmo IA (1980) Currents trends in the estimation of Michaelis-Menten parameters. Anal Biochem 104: 1–9 - PubMed
-
- Bachmann SL, McCarthy AJ (1989) Purification and characterization of a thermostable beta-xylosidase from Thermomonospora fusca. J Gen Microbiol 135: 293–299
-
- Banik M, Li CD, Langridge P, Fincher GB (1997) Structure, hormonal regulation, and chromosomal location of genes encoding barley (1-4)-beta-xylan endohydrolases. Mol Gen Genet 253: 599–608 - PubMed
-
- Beg QK, Kapoor M, Mahajan L, Hoondal GS (2001) Microbiol xylanases and their industrial applications: a review. Appl Microbiol Biotechnol 56: 326–338 - PubMed
-
- Bewley JD, Burton RA, Morohashi Y, Fincher GB (1997) Molecular cloning of a cDNA encoding a (1-4)-beta-mannan endohydrolase from the seeds of germinated tomato (Lycopersicon esculentum). Planta 203: 454–459 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

