Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life
- PMID: 15169916
- PMCID: PMC419860
- DOI: 10.1128/MCB.24.12.5577-5586.2004
Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life
Abstract
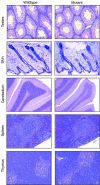
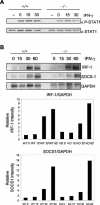
Protein inhibitor of activated STAT Y (PIASy) is the shortest member of the PIAS family and has been reported to modulate the transcriptional activities of STAT1, lymphoid enhancer factor 1 (LEF-1), and the androgen receptor. PIAS proteins have also been identified as E3 ligases for the small ubiquitin-like modifier (SUMO) proteins. PIASy in particular has been reported to mediate SUMO-2/3 modification of LEF-1, sequestering it into nuclear bodies, and SUMO-1 ligation to c-Myb, modulating its transcriptional activation properties. We have cloned murine Piasy and a splice variant which omits exon 6, containing the nuclear retention PINIT motif. Cell culture studies indicate that both the full length and the splice variant are localized in the nucleus but differentially enhance SUMO ligation. To further understand the functions of PIASy, we have generated PIASy-deficient mice. Surprisingly, Piasy(-/-) mice appear phenotypically normal. Activation of STAT1 is not significantly perturbed in Piasy(-/-) cells, and sumoylation patterns for SUMO-1 or SUMO-3 modification are similar when comparing tissues and embryonic fibroblasts from wild-type and knockout mice. Our study demonstrates that at steady state, PIASy is either dispensable or compensated for by other PIAS family members or by other mechanisms when deleted.
Figures





Similar articles
-
Identification of a new small ubiquitin-like modifier (SUMO)-interacting motif in the E3 ligase PIASy.J Biol Chem. 2017 Jun 16;292(24):10230-10238. doi: 10.1074/jbc.M117.789982. Epub 2017 Apr 28. J Biol Chem. 2017. PMID: 28455449 Free PMC article.
-
Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy.Circ Res. 2003 Jun 13;92(11):1201-8. doi: 10.1161/01.RES.0000076893.70898.36. Epub 2003 May 15. Circ Res. 2003. PMID: 12750312
-
PIASy-mediated repression of the androgen receptor is independent of sumoylation.Oncogene. 2004 Apr 15;23(17):3059-66. doi: 10.1038/sj.onc.1207443. Oncogene. 2004. PMID: 14981544
-
PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription.Biochem Soc Trans. 2007 Dec;35(Pt 6):1405-8. doi: 10.1042/BST0351405. Biochem Soc Trans. 2007. PMID: 18031232 Review.
-
A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases.Genes Dev. 2001 Dec 1;15(23):3053-8. doi: 10.1101/gad.955501. Genes Dev. 2001. PMID: 11731472 Review. No abstract available.
Cited by
-
PIASgamma is required for faithful chromosome segregation in human cells.PLoS One. 2006 Dec 20;1(1):e53. doi: 10.1371/journal.pone.0000053. PLoS One. 2006. PMID: 17183683 Free PMC article.
-
PIASy mediates SUMO-2/3 conjugation of poly(ADP-ribose) polymerase 1 (PARP1) on mitotic chromosomes.J Biol Chem. 2010 May 7;285(19):14415-23. doi: 10.1074/jbc.M109.074583. Epub 2010 Mar 12. J Biol Chem. 2010. PMID: 20228053 Free PMC article.
-
Identification of a new site of sumoylation on Tel (ETV6) uncovers a PIAS-dependent mode of regulating Tel function.Mol Cell Biol. 2008 Apr;28(7):2342-57. doi: 10.1128/MCB.01159-07. Epub 2008 Jan 22. Mol Cell Biol. 2008. PMID: 18212042 Free PMC article.
-
Spatial interplay between PIASy and FIP200 in the regulation of signal transduction and transcriptional activity.Mol Cell Biol. 2008 Apr;28(8):2771-81. doi: 10.1128/MCB.01210-07. Epub 2008 Feb 19. Mol Cell Biol. 2008. PMID: 18285457 Free PMC article.
-
SUMO: From Bench to Bedside.Physiol Rev. 2020 Oct 1;100(4):1599-1619. doi: 10.1152/physrev.00025.2019. Physiol Rev. 2020. PMID: 32666886 Free PMC article. Review.
References
-
- Chung, C. D., J. Liao, B. Liu, X. Rao, P. Jay, P. Berta, and K. Shuai. 1997. Specific inhibition of Stat3 signal transduction by PIAS3. Science 278:1803-1805. - PubMed
-
- Dahle, O., T. O. Andersen, O. Nordgard, V. Matre, G. Del Sal, and O. S. Gabrielsen. 2003. Transactivation properties of c-Myb are critically dependent on two SUMO-1 acceptor sites that are conjugated in a PIASy enhanced manner. Eur. J. Biochem. 270:1338-1348. - PubMed
-
- Dohmen, R. J., R. Stappen, J. P. McGrath, H. Forrova, J. Kolarov, A. Goffeau, and A. Varshavsky. 1995. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 270:18099-18109. - PubMed
-
- Duval, D., G. Duval, C. Kedinger, O. Poch, and H. Boeuf. 2003. The ′PINIT' motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 554:111-118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
