Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors
- PMID: 15158473
- PMCID: PMC7111185
- DOI: 10.1016/j.bbrc.2004.04.141
Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors
Abstract
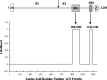
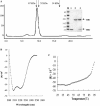
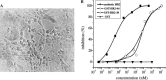
Severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) is a newly identified member of Family Coronaviridae. Coronavirus envelope spike protein S is a class I viral fusion protein which is characterized by the existence of two heptad repeat regions (HR1 and HR2) (forming a complex called fusion core). Here we report that by using in vitro bio-engineering techniques, SARS-CoV HR1 and HR2 bind to each other and form a typical 6-helix bundle. The HR2, either as a synthetic peptide or as a GST-fusion polypeptide, is a potent inhibitor of virus entry. The results do show that SARS-CoV follows the general fusion mechanism of class I viruses and this lays the ground for identification of virus fusion/entry inhibitors for this devastating emerging virus.
Figures
Similar articles
-
Characterization of the heptad repeat regions, HR1 and HR2, and design of a fusion core structure model of the spike protein from severe acute respiratory syndrome (SARS) coronavirus.Biochemistry. 2004 Nov 9;43(44):14064-71. doi: 10.1021/bi049101q. Biochemistry. 2004. PMID: 15518555
-
Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors.Lancet. 2004 Mar 20;363(9413):938-47. doi: 10.1016/S0140-6736(04)15788-7. Lancet. 2004. PMID: 15043961 Free PMC article.
-
Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core.J Biol Chem. 2004 Nov 19;279(47):49414-9. doi: 10.1074/jbc.M408782200. Epub 2004 Sep 1. J Biol Chem. 2004. PMID: 15345712 Free PMC article.
-
The SARS-CoV S glycoprotein.Cell Mol Life Sci. 2004 Oct;61(19-20):2428-30. doi: 10.1007/s00018-004-4257-y. Cell Mol Life Sci. 2004. PMID: 15526150 Free PMC article. Review.
-
Multimerization of the heptad repeat regions of the SARS-CoV 2 spike protein.Biochim Biophys Acta Biomembr. 2024 Feb;1866(2):184259. doi: 10.1016/j.bbamem.2023.184259. Epub 2023 Dec 5. Biochim Biophys Acta Biomembr. 2024. PMID: 38061554 Review.
Cited by
-
Druggable targets from coronaviruses for designing new antiviral drugs.Bioorg Med Chem. 2020 Nov 15;28(22):115745. doi: 10.1016/j.bmc.2020.115745. Epub 2020 Sep 8. Bioorg Med Chem. 2020. PMID: 33007557 Free PMC article. Review.
-
Design of recombinant protein-based SARS-CoV entry inhibitors targeting the heptad-repeat regions of the spike protein S2 domain.Biochem Biophys Res Commun. 2005 Apr 29;330(1):39-45. doi: 10.1016/j.bbrc.2005.02.117. Biochem Biophys Res Commun. 2005. PMID: 15781229 Free PMC article.
-
Towards our understanding of SARS-CoV, an emerging and devastating but quickly conquered virus.Comp Immunol Microbiol Infect Dis. 2007 Sep;30(5-6):309-27. doi: 10.1016/j.cimid.2007.05.009. Epub 2007 Jul 19. Comp Immunol Microbiol Infect Dis. 2007. PMID: 17640731 Free PMC article. Review.
-
Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus.J Virol. 2013 Dec;87(24):13134-40. doi: 10.1128/JVI.02433-13. Epub 2013 Sep 25. J Virol. 2013. PMID: 24067982 Free PMC article.
-
Characterization of neutralizing monoclonal antibodies recognizing a 15-residues epitope on the spike protein HR2 region of severe acute respiratory syndrome coronavirus (SARS-CoV).J Biomed Sci. 2005 Oct;12(5):711-27. doi: 10.1007/s11373-005-9004-3. Epub 2005 Nov 9. J Biomed Sci. 2005. PMID: 16132115 Free PMC article.
References
-
- Ksiazek T.G, Erdman D, Goldsmith C.S, Zaki S.R, Peret T, Emery S, Tong S, Urbani C, Comer J.A, Lim W, Rollin P.E, Dowell S.F, Ling A.E, Humphrey C.D, Shieh W.J, Guarner J, Paddock C.D, Rota P, Fields B, DeRisi J, Yang J.Y, Cox N, Hughes J.M, LeDuc J.W, Bellini W.J, Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
-
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt H.R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier R.A, Berger A, Burguiere A.M, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra J.C, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk H.D, Osterhaus A.D, Schmitz H, Doerr H.W. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Marra M.A, Jones S.J, Astell C.R, Holt R.A, Brooks-Wilson A, Butterfield Y.S, Khattra J, Asano J.K, Barber S.A, Chan S.Y, Cloutier A, Coughlin S.M, Freeman D, Girn N, Griffith O.L, Leach S.R, Mayo M, McDonald H, Montgomery S.B, Pandoh P.K, Petrescu A.S, Robertson A.G, Schein J.E, Siddiqui A, Smailus D.E, Stott J.M, Yang G.S, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth T.F, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples G.A, Tyler S, Vogrig R, Ward D, Watson B, Brunham R.C, Krajden M, Petric M, Skowronski D.M, Upton C, Roper R.L. Science. 2003;300:1399–1404. - PubMed
-
- Rota P.A, Oberste M.S, Monroe S.S, Nix W.A, Campagnoli R, Icenogle J.P, Penaranda S, Bankamp B, Maher K, Chen M.H, Tong S, Tamin A, Lowe L, Frace M, DeRisi J.L, Chen Q, Wang D, Erdman D.D, Peret T.C, Burns C, Ksiazek T.G, Rollin P.E, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus A.D, Drosten C, Pallansch M.A, Anderson L.J, Bellini W.J. Science. 2003;300:1394–1399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

