Protein-lipid interactions and phosphoinositide metabolism in membrane traffic: insights from vesicle recycling in nerve terminals
- PMID: 15146067
- PMCID: PMC420382
- DOI: 10.1073/pnas.0401874101
Protein-lipid interactions and phosphoinositide metabolism in membrane traffic: insights from vesicle recycling in nerve terminals
Abstract
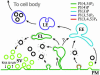
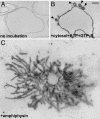
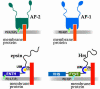
Great progress has been made in the elucidation of the function of proteins in membrane traffic. Less is known about the regulatory role of lipids in membrane dynamics. Studies of nerve terminals, compartments highly specialized for the recycling of synaptic vesicles, have converged with studies from other systems to reveal mechanisms in protein-lipid interactions that affect membrane shape as well as the fusion and fission of vesicles. Phosphoinositides have emerged as major regulators of the binding of cytosolic proteins to the bilayer. Phosphorylation on different positions of the inositol ring generates different isomers that are heterogeneously distributed on cell membranes and that together with membrane proteins generate a "dual keys" code for the recruitment of cytosolic proteins. This code helps controlling vectoriality of membrane transport. Powerful methods for the detection of lipids are rapidly advancing this field, thus complementing the broad range of information about biological systems that can be obtained from genomic and proteomic approaches.
Figures

Similar articles
-
Phosphoinositides in membrane traffic at the synapse.J Cell Sci. 2001 Mar;114(Pt 6):1041-52. doi: 10.1242/jcs.114.6.1041. J Cell Sci. 2001. PMID: 11228149 Review.
-
Mechanisms of synaptic vesicle recycling illuminated by fluorescent dyes.J Neurochem. 1999 Dec;73(6):2227-39. doi: 10.1046/j.1471-4159.1999.0732227.x. J Neurochem. 1999. PMID: 10582580 Review.
-
Phosphoinositides as spatial regulators of membrane traffic.Curr Opin Neurobiol. 1997 Jun;7(3):331-8. doi: 10.1016/s0959-4388(97)80060-8. Curr Opin Neurobiol. 1997. PMID: 9232806 Review.
-
The eighth Datta Lecture. Molecular mechanisms in synaptic vesicle recycling.FEBS Lett. 1995 Aug 1;369(1):3-12. doi: 10.1016/0014-5793(95)00739-v. FEBS Lett. 1995. PMID: 7641879 Review.
-
Syntaxin-1A is excluded from recycling synaptic vesicles at nerve terminals.J Neurosci. 2004 May 19;24(20):4884-8. doi: 10.1523/JNEUROSCI.0174-04.2004. J Neurosci. 2004. PMID: 15152049 Free PMC article.
Cited by
-
The physical influence of inositides-a disproportionate effect?J Chem Biol. 2014 Jul 20;8(1):1-3. doi: 10.1007/s12154-014-0117-x. eCollection 2015 Jan. J Chem Biol. 2014. PMID: 25584076 Free PMC article.
-
Coordinated Expression of Phosphoinositide Metabolic Genes during Development and Aging of Human Dorsolateral Prefrontal Cortex.PLoS One. 2015 Jul 13;10(7):e0132675. doi: 10.1371/journal.pone.0132675. eCollection 2015. PLoS One. 2015. PMID: 26168237 Free PMC article.
-
Connecdenn, a novel DENN domain-containing protein of neuronal clathrin-coated vesicles functioning in synaptic vesicle endocytosis.J Neurosci. 2006 Dec 20;26(51):13202-12. doi: 10.1523/JNEUROSCI.4608-06.2006. J Neurosci. 2006. PMID: 17182770 Free PMC article.
-
Palmitoylation controls the catalytic activity and subcellular distribution of phosphatidylinositol 4-kinase II{alpha}.J Biol Chem. 2009 Apr 10;284(15):9994-10003. doi: 10.1074/jbc.M900724200. Epub 2009 Feb 11. J Biol Chem. 2009. PMID: 19211550 Free PMC article.
-
Endosomal Phosphatidylinositol 3-Phosphate Promotes Gephyrin Clustering and GABAergic Neurotransmission at Inhibitory Postsynapses.J Biol Chem. 2017 Jan 27;292(4):1160-1177. doi: 10.1074/jbc.M116.771592. Epub 2016 Dec 9. J Biol Chem. 2017. PMID: 27941024 Free PMC article.
References
-
- Jahn, R., Lang, T. & Sudhof, T. C. (2003) Cell 112, 519-533. - PubMed
-
- Rothman, J. E. (2002) Nat. Med. 8, 1059-1062. - PubMed
-
- Israelachvili, J. N. & Mitchell, D. J. (1975) Biochim. Biophys. Acta 389, 13-19. - PubMed
-
- Chernomordik, L., Kozlov, M. M. & Zimmerberg, J. (1995) J. Membr. Biol. 146, 1-14. - PubMed
-
- Burger, K. N. (2000) Traffic 1, 605-613. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

