A plasmid-based system for expressing small interfering RNA libraries in mammalian cells
- PMID: 15119963
- PMCID: PMC416474
- DOI: 10.1186/1471-2121-5-16
A plasmid-based system for expressing small interfering RNA libraries in mammalian cells
Abstract
Background: RNA interference (RNAi) is an evolutionarily conserved process that functions to inhibit gene expression. The use of RNAi in mammals as a tool to study gene function has rapidly developed in the last couple of years since the discovery that the function-inhibiting units of RNAi are short 21-25 nt double-stranded RNAs (siRNAs) derived from their longer template. The use of siRNAs allows for gene-specific knock-down without induction of the non-specific interferon response in mammalian cells. Multiple systems have been developed to introduce siRNAs into mammals. One of the most appealing of these techniques is the use of vectors containing polymerase III promoters to drive expression of hairpin siRNAs. However, there are multiple limitations to using hairpin siRNA vectors including the observation that some are unstable in bacteria and are difficult to sequence.
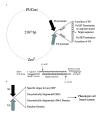
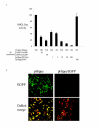
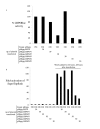
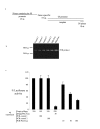
Results: To circumvent the limitation of hairpin siRNA vectors we have developed a convergent opposing siRNA expression system called pHippy. We have generated pHippy vectors or expression cassettes that knock down the expression of both reporter and endogenous genes. As a proof of principle that pHippy can be used to generate random siRNA libraries, we generated a small siRNA library against PGL3 luciferase and demonstrated that we could recover functional siRNAs that knock down PGL3 luciferase.
Conclusions: siRNA is a powerful tool to study gene function. We have developed a new vector with opposing convergent promoters for the expression of siRNAs, which can be used to knock down endogenous genes in a high throughput manner or to perform functional screening with random or cDNA-derived siRNA libraries.
Figures

Similar articles
-
Effective inhibition of hepatitis B virus replication by small interfering RNAs expressed from human foamy virus vectors.Int J Mol Med. 2007 Apr;19(4):705-11. Int J Mol Med. 2007. PMID: 17334648
-
Screening of siRNA target sequences by using fragmentized DNA.J Gene Med. 2006 Jun;8(6):782-91. doi: 10.1002/jgm.900. J Gene Med. 2006. PMID: 16532512
-
[An efficient method for screening effective siRNAs using dual-luciferase reporter assay system].Nan Fang Yi Ke Da Xue Xue Bao. 2009 Aug;29(8):1577-81. Nan Fang Yi Ke Da Xue Xue Bao. 2009. PMID: 19726297 Chinese.
-
Genome-wide application of RNAi to the discovery of potential drug targets.FEBS Lett. 2005 Oct 31;579(26):5988-95. doi: 10.1016/j.febslet.2005.08.015. Epub 2005 Aug 22. FEBS Lett. 2005. PMID: 16153642 Review.
-
Vectors for RNA interference.Curr Opin Mol Ther. 2004 Aug;6(4):367-72. Curr Opin Mol Ther. 2004. PMID: 15468595 Review.
Cited by
-
Genome-Wide RNAi Screening Identifies Novel Pathways/Genes Involved in Oxidative Stress and Repurposable Drugs to Preserve Cystic Fibrosis Airway Epithelial Cell Integrity.Antioxidants (Basel). 2021 Dec 2;10(12):1936. doi: 10.3390/antiox10121936. Antioxidants (Basel). 2021. PMID: 34943039 Free PMC article.
-
Identification of GAS1 as an epirubicin resistance-related gene in human gastric cancer cells with a partially randomized small interfering RNA library.J Biol Chem. 2009 Sep 25;284(39):26273-85. doi: 10.1074/jbc.M109.028068. Epub 2009 Jul 28. J Biol Chem. 2009. PMID: 19638344 Free PMC article.
-
Generation of shRNAs from randomized oligonucleotides.Biol Proced Online. 2007 Jul 4;9:9-17. doi: 10.1251/bpo129. Biol Proced Online. 2007. PMID: 18213360 Free PMC article.
-
RNA modulators of complex phenotypes in mammalian cells.PLoS One. 2009;4(3):e4758. doi: 10.1371/journal.pone.0004758. Epub 2009 Mar 9. PLoS One. 2009. PMID: 19270743 Free PMC article.
-
Dazap2 modulates transcription driven by the Wnt effector TCF-4.Nucleic Acids Res. 2009 May;37(9):3007-20. doi: 10.1093/nar/gkp179. Epub 2009 Mar 20. Nucleic Acids Res. 2009. PMID: 19304756 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

