Structure and mechanism of the RNA polymerase II transcription machinery
- PMID: 15114340
- PMCID: PMC1189732
- DOI: 10.1038/nsmb763
Structure and mechanism of the RNA polymerase II transcription machinery
Abstract
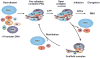
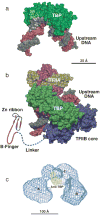
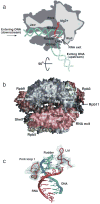
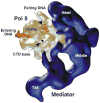
Advances in structure determination of the bacterial and eukaryotic transcription machinery have led to a marked increase in the understanding of the mechanism of transcription. Models for the specific assembly of the RNA polymerase II transcription machinery at a promoter, conformational changes that occur during initiation of transcription, and the mechanism of initiation are discussed in light of recent developments.
Conflict of interest statement
The author declares that he has no competing financial interests.
Figures

Similar articles
-
Structural basis of transcription: separation of RNA from DNA by RNA polymerase II.Science. 2004 Feb 13;303(5660):1014-6. doi: 10.1126/science.1090839. Science. 2004. PMID: 14963331
-
The RNA polymerase II core promoter.Annu Rev Biochem. 2003;72:449-79. doi: 10.1146/annurev.biochem.72.121801.161520. Epub 2003 Mar 19. Annu Rev Biochem. 2003. PMID: 12651739 Review.
-
Eukaryotic transcription initiation machinery visualized at molecular level.Transcription. 2016 Oct 19;7(5):203-208. doi: 10.1080/21541264.2016.1237150. Transcription. 2016. PMID: 27658022 Free PMC article. Review.
-
RNA polymerase II as a control panel for multiple coactivator complexes.Curr Opin Genet Dev. 1999 Apr;9(2):132-9. doi: 10.1016/S0959-437X(99)80020-3. Curr Opin Genet Dev. 1999. PMID: 10322136 Review.
-
Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms.Science. 2004 Feb 13;303(5660):983-8. doi: 10.1126/science.1090838. Science. 2004. PMID: 14963322
Cited by
-
Enzymatic Protein Biopolymers as a Tool to Synthetize Eukaryotic Messenger Ribonucleic Acid (mRNA) with Uses in Vaccination, Immunotherapy and Nanotechnology.Polymers (Basel). 2020 Jul 23;12(8):1633. doi: 10.3390/polym12081633. Polymers (Basel). 2020. PMID: 32717794 Free PMC article. Review.
-
Evidence that RNA polymerase II and not TFIIB is responsible for the difference in transcription initiation patterns between Saccharomyces cerevisiae and Schizosaccharomyces pombe.Nucleic Acids Res. 2012 Aug;40(14):6495-507. doi: 10.1093/nar/gks323. Epub 2012 Apr 17. Nucleic Acids Res. 2012. PMID: 22510268 Free PMC article.
-
Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules.Nat Commun. 2015 Jan 14;6:6011. doi: 10.1038/ncomms7011. Nat Commun. 2015. PMID: 25586196 Free PMC article.
-
Transcription and Maturation of mRNA in Dinoflagellates.Microorganisms. 2013 Nov 1;1(1):71-99. doi: 10.3390/microorganisms1010071. Microorganisms. 2013. PMID: 27694765 Free PMC article. Review.
-
Uncovering the mechanisms of transcription elongation by eukaryotic RNA polymerases I, II, and III.iScience. 2022 Oct 8;25(11):105306. doi: 10.1016/j.isci.2022.105306. eCollection 2022 Nov 18. iScience. 2022. PMID: 36304104 Free PMC article.
References
-
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. - PubMed
-
- Woychik NA, Hampsey M. The RNA polymerase II machinery: structure illuminates function. Cell. 2002;108:453–463. - PubMed
-
- Borukhov S, Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Curr Opin Microbiol. 2003;6:93–100. - PubMed
-
- Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

