A genomic selection strategy to identify accessible and dimerization blocking targets in the 5'-UTR of HIV-1 RNA
- PMID: 15107482
- PMCID: PMC407842
- DOI: 10.1093/nar/gnh064
A genomic selection strategy to identify accessible and dimerization blocking targets in the 5'-UTR of HIV-1 RNA
Abstract
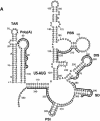
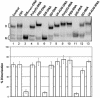
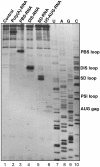
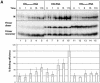
Defining target sites for antisense oligonucleotides in highly structured RNA is a non-trivial exercise that has received much attention. Here we describe a novel and simple method to generate a library composed of all 20mer oligoribonucleotides that are sense- and antisense to any given sequence or genome and apply the method to the highly structured HIV-1 leader RNA. Oligoribonucleotides that interact strongly with folded HIV-1 RNA and potentially inhibit its dimerization were identified through iterative rounds of affinity selection by native gel electrophoresis. We identified five distinct regions in the HIV-1 RNA that were particularly prone to antisense annealing and a structural comparison between these sites suggested that the 3'-end of the antisense RNA preferentially interacts with single-stranded loops in the target RNA, whereas the 5'-end binds within double-stranded regions. The selected RNA species and corresponding DNA oligonucleotides were assayed for HIV-1 RNA binding, ability to block reverse transcription and/or potential to interfere with dimerization. All the selected oligonucleotides bound rapidly and strongly to the HIV-1 leader RNA in vitro and one oligonucleotide was capable of disrupting RNA dimers efficiently. The library selection methodology we describe here is rapid, inexpensive and generally applicable to any other RNA or RNP complex. The length of the oligonucleotide in the library is similar to antisense molecules generally applied in vivo and therefore likely to define targets relevant for HIV-1 therapy.
Figures





Similar articles
-
Role of the 5' TAR stem--loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA.Biochemistry. 2008 Mar 11;47(10):3283-93. doi: 10.1021/bi7023173. Epub 2008 Feb 16. Biochemistry. 2008. PMID: 18278873
-
Selective binding of hepatitis C virus core protein to synthetic oligonucleotides corresponding to the 5' untranslated region of the viral genome.Virology. 2000 Apr 25;270(1):229-36. doi: 10.1006/viro.2000.0252. Virology. 2000. PMID: 10772995
-
Cooperative and specific binding of Vif to the 5' region of HIV-1 genomic RNA.J Mol Biol. 2005 Nov 18;354(1):55-72. doi: 10.1016/j.jmb.2005.09.025. Epub 2005 Oct 3. J Mol Biol. 2005. PMID: 16236319
-
Regulating eukaryotic gene expression with aptamers.FEBS Lett. 2004 Jun 1;567(1):55-62. doi: 10.1016/j.febslet.2004.03.111. FEBS Lett. 2004. PMID: 15165893 Review.
-
A structure-based approach for targeting the HIV-1 genomic RNA dimerization initiation site.Biochimie. 2007 Oct;89(10):1195-203. doi: 10.1016/j.biochi.2007.03.003. Epub 2007 Mar 12. Biochimie. 2007. PMID: 17434658 Review.
Cited by
-
Short-hairpin RNAs synthesized by T7 phage polymerase do not induce interferon.Nucleic Acids Res. 2008 Feb;36(3):e18. doi: 10.1093/nar/gkm1043. Epub 2008 Jan 21. Nucleic Acids Res. 2008. PMID: 18208841 Free PMC article.
-
HIV-1 RNA dimerization: It takes two to tango.AIDS Rev. 2009 Apr-Jun;11(2):91-102. AIDS Rev. 2009. PMID: 19529749 Free PMC article. Review.
-
Modelling toehold-mediated RNA strand displacement.Biophys J. 2015 Mar 10;108(5):1238-47. doi: 10.1016/j.bpj.2015.01.023. Biophys J. 2015. PMID: 25762335 Free PMC article.
-
Locked nucleic acid containing antisense oligonucleotides enhance inhibition of HIV-1 genome dimerization and inhibit virus replication.FEBS Lett. 2004 Dec 17;578(3):285-90. doi: 10.1016/j.febslet.2004.11.015. FEBS Lett. 2004. PMID: 15589834 Free PMC article.
-
XNAzymes targeting the SARS-CoV-2 genome inhibit viral infection.Nat Commun. 2022 Nov 16;13(1):6716. doi: 10.1038/s41467-022-34339-w. Nat Commun. 2022. PMID: 36385143 Free PMC article.
References
-
- Kurreck J. (2003) Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem., 270, 1628–1644. - PubMed
-
- Sohail M. and Southern,E.M. (2000) Selecting optimal antisense reagents. Adv. Drug Deliv. Rev., 44, 23–34. - PubMed
-
- Sohail M. and Southern,E.M. (2000) Hybridization of antisense reagents to RNA. Curr. Opin. Mol. Ther., 2, 264–271. - PubMed
-
- Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. - PubMed
-
- Bohula E.A., Salisbury,A.J., Sohail,M., Playford,M.P., Riedemann,J., Southern,E.M. and Macaulay,V.M. (2003) The efficacy of small interfering RNAs targeted to the type 1 insulin-like growth factor receptor (IGF1R) is influenced by secondary structure in the IGF1R transcript. J. Biol. Chem., 278, 15991–15997. - PubMed

