Molecular evolution and circulation patterns of human respiratory syncytial virus subgroup a: positively selected sites in the attachment g glycoprotein
- PMID: 15078950
- PMCID: PMC387670
- DOI: 10.1128/jvi.78.9.4675-4683.2004
Molecular evolution and circulation patterns of human respiratory syncytial virus subgroup a: positively selected sites in the attachment g glycoprotein
Abstract
Human respiratory syncytial virus (HRSV) is the most common etiological agent of acute lower respiratory tract disease in infants and can cause repeated infections throughout life. In this study, we have analyzed nucleotide sequences encompassing 629 bp at the carboxy terminus of the G glycoprotein gene for HRSV subgroup A strains isolated over 47 years, including 112 Belgian strains isolated over 19 consecutive years (1984 to 2002). By using a maximum likelihood method, we have tested the presence of diversifying selection and identified 13 positively selected sites with a posterior probability above 0.5. The sites under positive selection correspond to sites of O glycosylation or to amino acids that were previously described as monoclonal antibody-induced in vitro escape mutants. Our findings suggest that the evolution of subgroup A HRSV G glycoprotein is driven by immune pressure operating in certain codon positions located mainly in the second hypervariable region of the ectodomain. Phylogenetic analysis revealed the prolonged cocirculation of two subgroup A lineages among the Belgian population and the possible extinction of three other lineages. The evolutionary rate of HRSV subgroup A isolates was estimated to be 1.83 x 10(-3) nucleotide substitutions/site/year, projecting the most recent common ancestor back to the early 1940s.
Figures
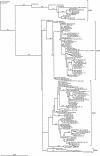
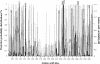
 ). Positively selected amino acid sites with posterior probabilities above 50% are indicated according to the Long strain. Amino acid variability, measured by entropy (Hi), is plotted to the second y axis.
). Positively selected amino acid sites with posterior probabilities above 50% are indicated according to the Long strain. Amino acid variability, measured by entropy (Hi), is plotted to the second y axis.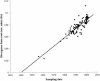
Similar articles
-
Molecular characterization of human respiratory syncytial virus subtype B: a novel genotype of subtype B circulating in China.J Med Virol. 2015 Jan;87(1):1-9. doi: 10.1002/jmv.23960. Epub 2014 Jun 9. J Med Virol. 2015. PMID: 24910250
-
Amino acid changes in the attachment G glycoprotein of human respiratory syncytial viruses (subgroup A) isolated in Italy over several epidemics (1997-2006).J Med Virol. 2007 Dec;79(12):1935-42. doi: 10.1002/jmv.21012. J Med Virol. 2007. PMID: 17935190
-
Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein.J Virol. 2005 Jul;79(14):9157-67. doi: 10.1128/JVI.79.14.9157-9167.2005. J Virol. 2005. PMID: 15994810 Free PMC article.
-
Molecular epidemiology of human respiratory syncytial virus in Uruguay: 1985-2001--a review.Mem Inst Oswaldo Cruz. 2005 May;100(3):221-30. doi: 10.1590/s0074-02762005000300001. Epub 2005 Aug 15. Mem Inst Oswaldo Cruz. 2005. PMID: 16113858 Review.
-
Characterization of human respiratory syncytial virus in children with severe acute respiratory infection before and during the COVID-19 pandemic.IJID Reg. 2024 Mar 21;11:100354. doi: 10.1016/j.ijregi.2024.03.009. eCollection 2024 Jun. IJID Reg. 2024. PMID: 38596821 Free PMC article. Review.
Cited by
-
Fully Bayesian tests of neutrality using genealogical summary statistics.BMC Genet. 2008 Oct 31;9:68. doi: 10.1186/1471-2156-9-68. BMC Genet. 2008. PMID: 18976476 Free PMC article.
-
Genetic diversity and evolutionary insights of respiratory syncytial virus A ON1 genotype: global and local transmission dynamics.Sci Rep. 2015 Sep 30;5:14268. doi: 10.1038/srep14268. Sci Rep. 2015. PMID: 26420660 Free PMC article.
-
Induction of IL-6 and CCL5 (RANTES) in human respiratory epithelial (A549) cells by clinical isolates of respiratory syncytial virus is strain specific.Virol J. 2012 Sep 10;9:190. doi: 10.1186/1743-422X-9-190. Virol J. 2012. PMID: 22962966 Free PMC article.
-
Positive natural selection in the evolution of human metapneumovirus attachment glycoprotein.Virus Res. 2008 Feb;131(2):121-31. doi: 10.1016/j.virusres.2007.08.014. Epub 2007 Oct 10. Virus Res. 2008. PMID: 17931731 Free PMC article.
-
Molecular characterization of circulating respiratory syncytial virus (RSV) genotypes in Gilgit Baltistan Province of Pakistan during 2011-2012 winter season.PLoS One. 2013 Sep 13;8(9):e74018. doi: 10.1371/journal.pone.0074018. eCollection 2013. PLoS One. 2013. PMID: 24058513 Free PMC article.
References
-
- Akerlind, B., and E. Norrby. 1986. Occurrence of respiratory syncytial virus subtypes A and B strains in Sweden. J. Med. Virol. 19:241-247. - PubMed
-
- Anderson, L. J., J. C. Hierholzer, C. Tsou, R. M. Hendry, B. F. Fernie, Y. Stone, and K. McIntosh. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626-633. - PubMed
-
- Cane, P. A. 2001. Molecular epidemiology of respiratory syncytial virus. Rev. Med. Virol. 11:103-116. - PubMed
-
- Cane, P. A. 1997. Analysis of linear epitopes recognised by the primary human antibody response to a variable region of the attachment (G) protein of respiratory syncytial virus. J. Med. Virol. 51:297-304. - PubMed
-
- Cane, P. A., and C. R. Pringle. 1995. Molecular epidemiology of respiratory syncytial virus: a review of the use of reverse transcription-polymerase chain reaction in the analysis of genetic variability. Electrophoresis 16:329-333. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

