The second RNA-binding domain of the human splicing factor ASF/SF2 is the critical domain controlling adenovirus E1A alternative 5'-splice site selection
- PMID: 15068396
- PMCID: PMC1133838
- DOI: 10.1042/BJ20040408
The second RNA-binding domain of the human splicing factor ASF/SF2 is the critical domain controlling adenovirus E1A alternative 5'-splice site selection
Abstract
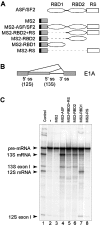
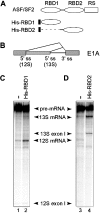
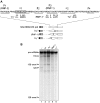
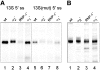
The human splicing factor ASF/SF2 (alternative splicing factor/splicing factor 2) is modular in structure with two RNA-binding domains (RBD1 and RBD2) and a C-terminal domain rich in arginine-serine dipeptide repeats. ASF/SF2 is an essential splicing factor that also functions as an important regulator of alternative splicing. In adenovirus E1A (early region 1A) alternative pre-mRNA splicing, ASF/SF2 functions as a strong inducer of proximal 5'-splice-site selection, both in vitro and in vivo. In the present study, we tested the functional role of individual domains of ASF/SF2 in alternative splicing in vitro. We show that ASF/SF2-RBD2 is the critical domain controlling E1A alternative splicing. In fact, RBD2 alone is sufficient to mimic the activity of the full-length ASF/SF2 protein as an inducer of proximal 5'-splice-site selection in vitro. The RBD2 domain induces a switch to E1A-proximal 5'-splice-site usage by repressing distal 12 S splicing and simultaneously stimulates proximal 13 S splicing. In contrast, the ASF/SF2-RBD1 domain has a more general splicing enhancer phenotype and appears to stimulate preferentially cap-proximal 5'-splice-site selection. Furthermore, the SWQDLKD motif, which is conserved in all SR proteins (serine/arginine-rich proteins) containing two RBDs, and the ribonucleoprotein-1-type RNA recognition motif were both found to be necessary for the alternative splice-site-switching activity of ASF/SF2. The RNP-1 motif was necessary for efficient RNA binding, whereas the SWQDLKD motif most probably contributes by functioning as a surface-mediating critical protein-protein contact during spliceosome assembly.
Figures
Similar articles
-
Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing.J Biol Chem. 2002 Apr 12;277(15):12579-86. doi: 10.1074/jbc.M107867200. Epub 2002 Jan 18. J Biol Chem. 2002. PMID: 11801589
-
ASF/SF2-like maize pre-mRNA splicing factors affect splice site utilization and their transcripts are alternatively spliced.Gene. 2004 Sep 15;339:25-37. doi: 10.1016/j.gene.2004.06.047. Gene. 2004. PMID: 15363843
-
Exonic splicing enhancer-dependent selection of the bovine papillomavirus type 1 nucleotide 3225 3' splice site can be rescued in a cell lacking splicing factor ASF/SF2 through activation of the phosphatidylinositol 3-kinase/Akt pathway.J Virol. 2003 Feb;77(3):2105-15. doi: 10.1128/jvi.77.3.2105-2115.2003. J Virol. 2003. PMID: 12525645 Free PMC article.
-
Alternative splicing of pre-messenger RNAs in plants in the genomic era.Annu Rev Plant Biol. 2007;58:267-94. doi: 10.1146/annurev.arplant.58.032806.103754. Annu Rev Plant Biol. 2007. PMID: 17222076 Review.
-
Pre-mRNA splicing in the new millennium.Curr Opin Cell Biol. 2001 Jun;13(3):302-9. doi: 10.1016/s0955-0674(00)00212-x. Curr Opin Cell Biol. 2001. PMID: 11343900 Review.
Cited by
-
RNA recognition motif 2 directs the recruitment of SF2/ASF to nuclear stress bodies.Nucleic Acids Res. 2004 Aug 9;32(14):4127-36. doi: 10.1093/nar/gkh759. Print 2004. Nucleic Acids Res. 2004. PMID: 15302913 Free PMC article.
-
Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition.Proc Natl Acad Sci U S A. 2013 Jul 23;110(30):E2802-11. doi: 10.1073/pnas.1303445110. Epub 2013 Jul 8. Proc Natl Acad Sci U S A. 2013. PMID: 23836656 Free PMC article.
-
Virus usurps alternative splicing to clear the decks for infection.Virol J. 2023 Jun 20;20(1):131. doi: 10.1186/s12985-023-02098-9. Virol J. 2023. PMID: 37340420 Free PMC article. Review.
-
A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1.Mol Cell. 2008 Mar 14;29(5):563-76. doi: 10.1016/j.molcel.2007.12.017. Mol Cell. 2008. PMID: 18342604 Free PMC article.
-
Novel Splicing of Immune System Thyroid Stimulating Hormone β-Subunit-Genetic Regulation and Biological Importance.Front Endocrinol (Lausanne). 2019 Feb 5;10:44. doi: 10.3389/fendo.2019.00044. eCollection 2019. Front Endocrinol (Lausanne). 2019. PMID: 30804891 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

