The human beta-globin replication initiation region consists of two modular independent replicators
- PMID: 15060158
- PMCID: PMC381644
- DOI: 10.1128/MCB.24.8.3373-3386.2004
The human beta-globin replication initiation region consists of two modular independent replicators
Abstract
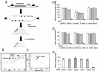
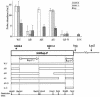
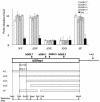
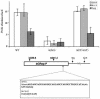
Previous studies have shown that mammalian cells contain replicator sequences, which can determine where DNA replication initiates. However, the specific sequences that confer replicator activity were not identified. Here we report a detailed analysis of replicator sequences that dictate initiation of DNA replication from the human beta-globin locus. This analysis suggests that the beta-globin replication initiation region contains two adjacent, redundant replicators. Each replicator was capable of initiating DNA replication independently at ectopic sites. Within each of these two replicators, we identified short, discrete, nonredundant sequences, which cooperatively determine replicator activity. Experiments with somatic cell hybrids further demonstrated that the requirements for initiation at ectopic sites were similar to the requirements for initiation within native human chromosomes. The replicator clustering and redundancy exemplified in the human beta-globin locus may account for the extreme difficulty in identifying replicator sequences in mammalian cells and suggest that mammalian replication initiation sites may be determined by cooperative sequence modules.
Figures




Similar articles
-
Cooperative sequence modules determine replication initiation sites at the human beta-globin locus.Hum Mol Genet. 2006 Sep 1;15(17):2613-22. doi: 10.1093/hmg/ddl187. Epub 2006 Jul 28. Hum Mol Genet. 2006. PMID: 16877501
-
Initiation of DNA replication at the human beta-globin 3' enhancer.Nucleic Acids Res. 2005 Aug 5;33(14):4412-24. doi: 10.1093/nar/gki747. Print 2005. Nucleic Acids Res. 2005. PMID: 16085752 Free PMC article.
-
The mammalian beta globin origin of DNA replication.Front Biosci. 2004 Sep 1;9:2540-7. doi: 10.2741/1415. Front Biosci. 2004. PMID: 15358579 Review.
-
Genetic dissection of a mammalian replicator in the human beta-globin locus.Science. 1998 Aug 14;281(5379):1005-9. doi: 10.1126/science.281.5379.1005. Science. 1998. PMID: 9703500
-
The replicon revisited: an old model learns new tricks in metazoan chromosomes.EMBO Rep. 2004 Jul;5(7):686-91. doi: 10.1038/sj.embor.7400185. EMBO Rep. 2004. PMID: 15229645 Free PMC article. Review.
Cited by
-
A winding road to origin discovery.Chromosome Res. 2010 Jan;18(1):45-61. doi: 10.1007/s10577-009-9089-z. Chromosome Res. 2010. PMID: 19859818 Free PMC article.
-
Binding of AlF-C, an Orc1-binding transcriptional regulator, enhances replicator activity of the rat aldolase B origin.Mol Cell Biol. 2006 Dec;26(23):8770-80. doi: 10.1128/MCB.00949-06. Epub 2006 Sep 18. Mol Cell Biol. 2006. PMID: 16982680 Free PMC article.
-
The hunt for origins of DNA replication in multicellular eukaryotes.F1000Prime Rep. 2015 Mar 3;7:30. doi: 10.12703/P7-30. eCollection 2015. F1000Prime Rep. 2015. PMID: 25926981 Free PMC article. Review.
-
DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin.Genes Dev. 2008 May 15;22(10):1319-24. doi: 10.1101/gad.468308. Epub 2008 Apr 28. Genes Dev. 2008. PMID: 18443145 Free PMC article.
-
The genetic architecture of DNA replication timing in human pluripotent stem cells.Nat Commun. 2021 Nov 19;12(1):6746. doi: 10.1038/s41467-021-27115-9. Nat Commun. 2021. PMID: 34799581 Free PMC article.
References
-
- Aladjem, M., L.-W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005-1009. - PubMed
-
- Aladjem, M., and G. M. Wahl. 1997. Mapping replication origins by leading strand analysis in the absence of protein synthesis. Methods Companion Methods Enzymol. 13:281-292. - PubMed
-
- Aladjem, M. I., M. Groudine, L. L. Brody, E. S. Dieken, R. E. K. Fournier, G. M. Wahl, and E. M. Epner. 1995. Participation of the human beta globin locus control region in initiation of DNA replication. Science 270:815-819. - PubMed
-
- Aladjem, M. I., L. W. Rodewald, C. M. Lin, S. Bowman, D. M. Cimbora, L. L. Brody, E. M. Epner, M. Groudine, and G. M. Wahl. 2002. Replication initiation patterns in the beta-globin loci of totipotent and differentiated murine cells: evidence for multiple initiation regions. Mol. Cell. Biol. 22:442-452. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
