Whole-genome DNA array analysis of the response of Borrelia burgdorferi to a bactericidal monoclonal antibody
- PMID: 15039324
- PMCID: PMC375205
- DOI: 10.1128/IAI.72.4.2035-2044.2004
Whole-genome DNA array analysis of the response of Borrelia burgdorferi to a bactericidal monoclonal antibody
Abstract
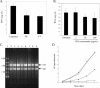
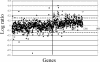
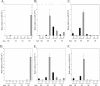
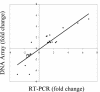
Identification and characterization of genes that contribute to infection with Borrelia burgdorferi and, of those, genes that are targets of host responses is important for understanding the pathogenesis of Lyme disease. The complement-independent bactericidal monoclonal antibody (MAb) CB2 recognizes a carboxy-terminal, hydrophilic epitope of the outer surface protein B (OspB). CB2 kills B. burgdorferi by an unknown bactericidal mechanism. Upon binding of CB2 to OspB, differentially expressed gene products may be responsible for, or associated with, the death of the organism. A time course of the response of B. burgdorferi to CB2 was completed to analyze the differential gene expression in the bacteria over a period of visual morphological changes. Bacteria were treated with a sublethal concentration in which spirochetes were visibly distressed by the antibody but not lysed. Preliminary whole-genome DNA arrays at various time points within 1 h of incubation of B. burgdorferi with the antibody showed that most significant changes occurred at 25 min. Circular plasmid 32 (cp32)-encoded genes were active in this period of time, including the blyA homologs, phage holin system genes. DNA array data show that three blyA homologs were upregulated significantly, >/==" BORDER="0">2 standard deviations from the mean of the log ratios, and a P value of </=0.01. Quantitative real-time PCR analysis verified blyA and blyB upregulation over an 18- to 35-min time course. The hypothesis to test is whether the killing mechanism of CB2 is through uncontrolled expression of the blyA and blyB phage holin system.
Figures

Similar articles
-
Cholesterol lipids of Borrelia burgdorferi form lipid rafts and are required for the bactericidal activity of a complement-independent antibody.Cell Host Microbe. 2010 Oct 21;8(4):331-42. doi: 10.1016/j.chom.2010.09.001. Cell Host Microbe. 2010. PMID: 20951967 Free PMC article.
-
Characterization of Borrelia burgdorferi BlyA and BlyB proteins: a prophage-encoded holin-like system.J Bacteriol. 2000 Dec;182(23):6791-7. doi: 10.1128/JB.182.23.6791-6797.2000. J Bacteriol. 2000. PMID: 11073925 Free PMC article.
-
Functional heterogeneity in the antibodies produced to Borrelia burgdorferi.Wien Klin Wochenschr. 1999 Dec 10;111(22-23):985-9. Wien Klin Wochenschr. 1999. PMID: 10666815 Review.
-
Characterization of the physiological requirements for the bactericidal effects of a monoclonal antibody to OspB of Borrelia burgdorferi by confocal microscopy.Infect Immun. 1997 May;65(5):1908-15. doi: 10.1128/iai.65.5.1908-1915.1997. Infect Immun. 1997. PMID: 9125579 Free PMC article.
-
Immune evasion of Borrelia burgdorferi: insufficient killing of the pathogens by complement and antibody.Int J Med Microbiol. 2002 Jun;291 Suppl 33:141-6. doi: 10.1016/s1438-4221(02)80027-3. Int J Med Microbiol. 2002. PMID: 12141738 Review.
Cited by
-
B1b lymphocyte-derived antibodies control Borrelia hermsii independent of Fcα/μ receptor and in the absence of host cell contact.Immunol Res. 2011 Dec;51(2-3):249-56. doi: 10.1007/s12026-011-8260-8. Immunol Res. 2011. PMID: 22139824 Free PMC article. Review.
-
Microarray-Based Comparative Genomic and Transcriptome Analysis of Borrelia burgdorferi.Microarrays (Basel). 2016 Apr 16;5(2):9. doi: 10.3390/microarrays5020009. Microarrays (Basel). 2016. PMID: 27600075 Free PMC article. Review.
-
Comparative transcriptional profiling of Borrelia burgdorferi clinical isolates differing in capacities for hematogenous dissemination.Infect Immun. 2005 Oct;73(10):6791-802. doi: 10.1128/IAI.73.10.6791-6802.2005. Infect Immun. 2005. PMID: 16177357 Free PMC article.
-
Oligopeptide permease A5 modulates vertebrate host-specific adaptation of Borrelia burgdorferi.Infect Immun. 2011 Aug;79(8):3407-20. doi: 10.1128/IAI.05234-11. Epub 2011 May 31. Infect Immun. 2011. PMID: 21628523 Free PMC article.
-
Overexpression of CsrA (BB0184) alters the morphology and antigen profiles of Borrelia burgdorferi.Infect Immun. 2009 Nov;77(11):5149-62. doi: 10.1128/IAI.00673-09. Epub 2009 Sep 8. Infect Immun. 2009. PMID: 19737901 Free PMC article.
References
-
- Anonymous. 2001. Lyme disease-United States, 1999. Morb. Mortal. Wkly. Rep. 50:181-185. - PubMed
-
- Arfin, S. M., A. D. Long, E. T. Ito, L. Tolleri, M. M. Riehle, E. S. Paegle, and G. W. Hatfield. 2000. Global gene expression profiling in Escherichia coli K-12: the effects of integration host factor. J. Biol. Chem. 275:29672-29684. - PubMed
-
- Baracchini, E., and H. Bremer. 1987. Determination of synthesis rate and lifetime of bacterial mRNAs. Anal. Biochem. 167:245-260. - PubMed
-
- Benach, J. L., E. M. Bosler, J. P. Hanrahan, J. L. Coleman, G. S. Habicht, T. F. Bast, D. J. Cameron, J. L. Ziegler, A. G. Barbour, W. Burgdorfer, R. Edelman, and R. A. Kaslow. 1983. Spirochetes isolated from the blood of two patients with Lyme disease. N. Engl. J. Med. 308:740-742. - PubMed
-
- Bergstrom, S., V. G. Bundoc, and A. G. Barbour. 1989. Molecular analysis of linear plasmid-encoded major surface proteins, OspA and OspB, of the Lyme disease spirochaete Borrelia burgdorferi. Mol. Microbiol. 3:479-486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

