Ubiquitin interactions of NZF zinc fingers
- PMID: 15029239
- PMCID: PMC391057
- DOI: 10.1038/sj.emboj.7600114
Ubiquitin interactions of NZF zinc fingers
Abstract
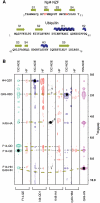
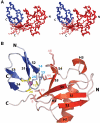
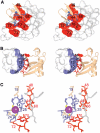
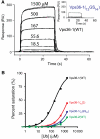
Ubiquitin (Ub) functions in many different biological pathways, where it typically interacts with proteins that contain modular Ub recognition domains. One such recognition domain is the Npl4 zinc finger (NZF), a compact zinc-binding module found in many proteins that function in Ub-dependent processes. We now report the solution structure of the NZF domain from Npl4 in complex with Ub. The structure reveals that three key NZF residues (13TF14/M25) surrounding the zinc coordination site bind the hydrophobic 'Ile44' surface of Ub. Mutations in the 13TF14/M25 motif inhibit Ub binding, and naturally occurring NZF domains that lack the motif do not bind Ub. However, substitution of the 13TF14/M25 motif into the nonbinding NZF domain from RanBP2 creates Ub-binding activity, demonstrating the versatility of the NZF scaffold. Finally, NZF mutations that inhibit Ub binding by the NZF domain of Vps36/ESCRT-II also inhibit sorting of ubiquitylated proteins into the yeast vacuole. Thus, the NZF is a versatile protein recognition domain that is used to bind ubiquitylated proteins during vacuolar protein sorting, and probably many other biological processes.
Figures

Similar articles
-
Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4.J Biol Chem. 2003 May 30;278(22):20225-34. doi: 10.1074/jbc.M300459200. Epub 2003 Mar 18. J Biol Chem. 2003. PMID: 12644454 Free PMC article.
-
Solution structure of the HOIL-1L NZF domain reveals a conformational switch regulating linear ubiquitin affinity.J Biol Chem. 2023 Sep;299(9):105165. doi: 10.1016/j.jbc.2023.105165. Epub 2023 Aug 16. J Biol Chem. 2023. PMID: 37595872 Free PMC article.
-
Dynamic recognition and linkage specificity in K63 di-ubiquitin and TAB2 NZF domain complex.Sci Rep. 2018 Nov 7;8(1):16478. doi: 10.1038/s41598-018-34605-2. Sci Rep. 2018. PMID: 30405169 Free PMC article.
-
Structural insights into specificity and diversity in mechanisms of ubiquitin recognition by ubiquitin-binding domains.Biochem Soc Trans. 2012 Apr;40(2):404-8. doi: 10.1042/BST20110729. Biochem Soc Trans. 2012. PMID: 22435820 Review.
-
When ubiquitin meets ubiquitin receptors: a signalling connection.Nat Rev Mol Cell Biol. 2003 Jun;4(6):491-7. doi: 10.1038/nrm1124. Nat Rev Mol Cell Biol. 2003. PMID: 12778128 Review.
Cited by
-
Molecular mechanisms of the inhibitory effects of bovine lactoferrin on lipopolysaccharide-mediated osteoclastogenesis.J Biol Chem. 2012 Jul 6;287(28):23527-36. doi: 10.1074/jbc.M111.324673. Epub 2012 May 16. J Biol Chem. 2012. PMID: 22593578 Free PMC article.
-
Peptide conformer acidity analysis of protein flexibility monitored by hydrogen exchange.Biochemistry. 2009 Oct 6;48(39):9256-65. doi: 10.1021/bi901219x. Biochemistry. 2009. PMID: 19722680 Free PMC article.
-
Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles.Mol Biol Cell. 2010 Mar 15;21(6):1023-32. doi: 10.1091/mbc.e09-09-0776. Epub 2010 Jan 20. Mol Biol Cell. 2010. PMID: 20089837 Free PMC article.
-
Solution structure of the C4 zinc finger domain of HDM2.Protein Sci. 2006 Feb;15(2):384-9. doi: 10.1110/ps.051927306. Epub 2005 Dec 29. Protein Sci. 2006. PMID: 16385008 Free PMC article.
-
Structural basis for the simultaneous recognition of NEMO and acceptor ubiquitin by the HOIP NZF1 domain.Sci Rep. 2022 Jul 18;12(1):12241. doi: 10.1038/s41598-022-16193-4. Sci Rep. 2022. PMID: 35851409 Free PMC article.
References
-
- Babst M, Katzmann D, Snyder W, Wendland B, Emr S (2002) Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 3: 283–289 - PubMed
-
- Bax A, Delaglio F, Grzesiek S, Vuister GW (1994a) Resonance assignment of methionine methyl groups and c3 angular information from long-range proton–carbon and carbon–carbon J correlation in a calmodulin–peptide complex. J Biomol NMR 4: 787–797 - PubMed
-
- Bax A, Vuister GW, Grzesiek S, Delaglio F, Wang AC, Tschudin R, Zhu G (1994b) Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol 239: 79–105 - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54 (Part 5): 905–921 - PubMed
-
- Buchberger A (2002) From UBA to UBX: new words in the ubiquitin vocabulary. Trends Cell Biol 12: 216–221 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

