Molecular imaging of drug-modulated protein-protein interactions in living subjects
- PMID: 15026351
- PMCID: PMC4154786
- DOI: 10.1158/0008-5472.can-03-2972
Molecular imaging of drug-modulated protein-protein interactions in living subjects
Abstract
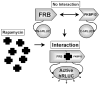
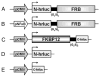
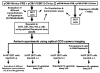
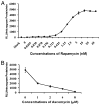
Networks of protein interactions mediate cellular responses to environmental stimuli and direct the execution of many different cellular functional pathways. Small molecules synthesized within cells or recruited from the external environment mediate many protein interactions. The study of small molecule-mediated interactions of proteins is important to understand abnormal signal transduction pathways in cancer and in drug development and validation. In this study, we used split synthetic renilla luciferase (hRLUC) protein fragment-assisted complementation to evaluate heterodimerization of the human proteins FRB and FKBP12 mediated by the small molecule rapamycin. The concentration of rapamycin required for efficient dimerization and that of its competitive binder ascomycin required for dimerization inhibition were studied in cell lines. The system was dually modulated in cell culture at the transcription level, by controlling nuclear factor kappaB promoter/enhancer elements using tumor necrosis factor alpha, and at the interaction level, by controlling the concentration of the dimerizer rapamycin. The rapamycin-mediated dimerization of FRB and FKBP12 also was studied in living mice by locating, quantifying, and timing the hRLUC complementation-based bioluminescence imaging signal using a cooled charged coupled device camera. This split reporter system can be used to efficiently screen small molecule drugs that modulate protein-protein interactions and also to assess drugs in living animals. Both are essential steps in the preclinical evaluation of candidate pharmaceutical agents targeting protein-protein interactions, including signaling pathways in cancer cells.
Figures

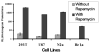


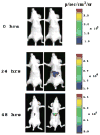

Similar articles
-
Novel fusion protein approach for efficient high-throughput screening of small molecule-mediating protein-protein interactions in cells and living animals.Cancer Res. 2005 Aug 15;65(16):7413-20. doi: 10.1158/0008-5472.CAN-05-0588. Cancer Res. 2005. PMID: 16103094 Free PMC article.
-
Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals.Proc Natl Acad Sci U S A. 2004 Aug 17;101(33):12288-93. doi: 10.1073/pnas.0404041101. Epub 2004 Jul 29. Proc Natl Acad Sci U S A. 2004. PMID: 15284440 Free PMC article.
-
Noninvasive imaging of protein-protein interactions from live cells and living subjects using bioluminescence resonance energy transfer.FASEB J. 2005 Dec;19(14):2017-9. doi: 10.1096/fj.05-4628fje. Epub 2005 Oct 4. FASEB J. 2005. PMID: 16204354
-
FK506-binding protein 12 ligands: a patent review.Expert Opin Ther Pat. 2013 Nov;23(11):1435-49. doi: 10.1517/13543776.2013.828695. Epub 2013 Aug 19. Expert Opin Ther Pat. 2013. PMID: 23957229 Review.
-
Chemical biology beyond binary codes.Chem Biol. 2000 Dec;7(12):R217-21. doi: 10.1016/s1074-5521(00)00040-5. Chem Biol. 2000. PMID: 11137822 Review.
Cited by
-
Molecular imaging of the efficacy of heat shock protein 90 inhibitors in living subjects.Cancer Res. 2008 Jan 1;68(1):216-26. doi: 10.1158/0008-5472.CAN-07-2268. Cancer Res. 2008. PMID: 18172314 Free PMC article.
-
Application of a split luciferase complementation assay for the detection of viral protein-protein interactions.J Virol Methods. 2011 Sep;176(1-2):108-11. doi: 10.1016/j.jviromet.2011.04.028. Epub 2011 May 30. J Virol Methods. 2011. PMID: 21645548 Free PMC article.
-
Bioluminescence: a versatile technique for imaging cellular and molecular features.Medchemcomm. 2014 Mar 1;5(3):255-267. doi: 10.1039/C3MD00288H. Epub 2013 Dec 13. Medchemcomm. 2014. PMID: 27594981 Free PMC article.
-
Firefly luciferase enzyme fragment complementation for imaging in cells and living animals.Anal Chem. 2005 Mar 1;77(5):1295-302. doi: 10.1021/ac0484777. Anal Chem. 2005. PMID: 15732910 Free PMC article.
-
Reporter gene imaging of protein-protein interactions in living subjects.Curr Opin Biotechnol. 2007 Feb;18(1):31-7. doi: 10.1016/j.copbio.2007.01.007. Epub 2007 Jan 24. Curr Opin Biotechnol. 2007. PMID: 17254764 Free PMC article. Review.
References
-
- Michnick SW. Exploring protein interactions by interaction-induced folding of proteins from complementary peptide fragments. Curr Opin Struct Biol. 2001;11:472–7. - PubMed
-
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–6. - PubMed
-
- Williams NE. Immunoprecipitation procedures. Methods Cell Biol. 2000;62:449–53. - PubMed
-
- Bollag DM. Gel-filtration chromatography. Methods Mol Biol. 1994;36:1–9. - PubMed
-
- Hansen JC, Lebowitz J, Demeler B. Analytical ultracentrifugation of complex macromolecular systems. Biochemistry. 1994;33:13155– 63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

