Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids
- PMID: 14981098
- PMCID: PMC2172170
- DOI: 10.1083/jcb.200308012
Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids
Abstract
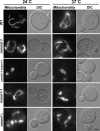
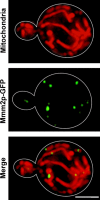
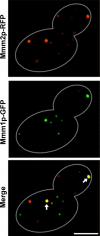
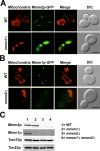
The mitochondrial outer membrane protein, Mmm1p, is required for normal mitochondrial shape in yeast. To identify new morphology proteins, we isolated mutations incompatible with the mmm1-1 mutant. One of these mutants, mmm2-1, is defective in a novel outer membrane protein. Lack of Mmm2p causes a defect in mitochondrial shape and loss of mitochondrial DNA (mtDNA) nucleoids. Like the Mmm1 protein (Aiken Hobbs, A.E., M. Srinivasan, J.M. McCaffery, and R.E. Jensen. 2001. J. Cell Biol. 152:401-410.), Mmm2p is located in dot-like particles on the mitochondrial surface, many of which are adjacent to mtDNA nucleoids. While some of the Mmm2p-containing spots colocalize with those containing Mmm1p, at least some of Mmm2p is separate from Mmm1p. Moreover, while Mmm2p and Mmm1p both appear to be part of large complexes, we find that Mmm2p and Mmm1p do not stably interact and appear to be members of two different structures. We speculate that Mmm2p and Mmm1p are components of independent machinery, whose dynamic interactions are required to maintain mitochondrial shape and mtDNA structure.
Figures



Similar articles
-
A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery.Mol Biol Cell. 2003 Nov;14(11):4618-27. doi: 10.1091/mbc.e03-04-0225. Epub 2003 Sep 17. Mol Biol Cell. 2003. PMID: 13679517 Free PMC article.
-
Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast.J Cell Biol. 2005 Jan 3;168(1):103-15. doi: 10.1083/jcb.200410030. J Cell Biol. 2005. PMID: 15631992 Free PMC article.
-
Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability.J Cell Biol. 2001 Jan 22;152(2):401-10. doi: 10.1083/jcb.152.2.401. J Cell Biol. 2001. PMID: 11266455 Free PMC article.
-
The enigmatic role of Mim1 in mitochondrial biogenesis.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):212-5. doi: 10.1016/j.ejcb.2009.11.002. Epub 2009 Nov 26. Eur J Cell Biol. 2010. PMID: 19944477 Review.
-
Organization and dynamics of yeast mitochondrial nucleoids.Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(5):339-359. doi: 10.2183/pjab.93.021. Proc Jpn Acad Ser B Phys Biol Sci. 2017. PMID: 28496055 Free PMC article. Review.
Cited by
-
Separating mitochondrial protein assembly and endoplasmic reticulum tethering by selective coupling of Mdm10.Nat Commun. 2016 Oct 10;7:13021. doi: 10.1038/ncomms13021. Nat Commun. 2016. PMID: 27721450 Free PMC article.
-
ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13.J Cell Biol. 2015 Sep 14;210(6):883-90. doi: 10.1083/jcb.201502105. J Cell Biol. 2015. PMID: 26370498 Free PMC article.
-
One ring to bring them all and in the darkness bind them: The trafficking of heme without deliverers.Biochim Biophys Acta Mol Cell Res. 2021 Jan;1868(1):118881. doi: 10.1016/j.bbamcr.2020.118881. Epub 2020 Oct 3. Biochim Biophys Acta Mol Cell Res. 2021. PMID: 33022276 Free PMC article. Review.
-
Lipid synthesis and membrane contact sites: a crossroads for cellular physiology.J Lipid Res. 2016 Oct;57(10):1789-1805. doi: 10.1194/jlr.R070920. Epub 2016 Aug 12. J Lipid Res. 2016. PMID: 27521373 Free PMC article. Review.
-
In Candida glabrata, ERMES Component GEM1 Controls Mitochondrial Morphology, mtROS, and Drug Efflux Pump Expression, Resulting in Azole Susceptibility.J Fungi (Basel). 2023 Feb 10;9(2):240. doi: 10.3390/jof9020240. J Fungi (Basel). 2023. PMID: 36836353 Free PMC article.
References
-
- Adams, A., D. Gottschling, C. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 177 pp.
-
- Attardi, G., and G. Schatz. 1988. Biogenesis of mitochondria. Annu. Rev. Cell Biol. 4:289–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

