Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression
- PMID: 14979921
- PMCID: PMC400653
- DOI: 10.1186/bcr754
Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression
Abstract
Introduction: In order to gain a better understanding of the molecular processes that underlie apoptosis and tissue regression in mammary gland, we undertook a large-scale analysis of transcriptional changes during the mouse mammary pregnancy cycle, with emphasis on the transition from lactation to involution.
Method: Affymetrix microarrays, representing 8618 genes, were used to compare mammary tissue from 12 time points (one virgin, three gestation, three lactation and five involution stages). Six animals were used for each time point. Common patterns of gene expression across all time points were identified and related to biological function.
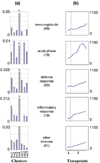
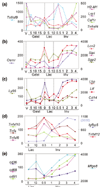
Results: The majority of significantly induced genes in involution were also differentially regulated at earlier stages in the pregnancy cycle. This included a marked increase in inflammatory mediators during involution and at parturition, which correlated with leukaemia inhibitory factor-Stat3 (signal transducer and activator of signalling-3) signalling. Before involution, expected increases in cell proliferation, biosynthesis and metabolism-related genes were observed. During involution, the first 24 hours after weaning was characterized by a transient increase in expression of components of the death receptor pathways of apoptosis, inflammatory cytokines and acute phase response genes. After 24 hours, regulators of intrinsic apoptosis were induced in conjunction with markers of phagocyte activity, matrix proteases, suppressors of neutrophils and soluble components of specific and innate immunity.
Conclusion: We provide a resource of mouse mammary gene expression data for download or online analysis. Here we highlight the sequential induction of distinct apoptosis pathways in involution and the stimulation of immunomodulatory signals, which probably suppress the potentially damaging effects of a cellular inflammatory response while maintaining an appropriate antimicrobial and phagocytic environment.
Figures
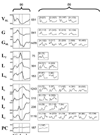
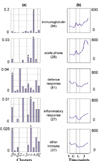
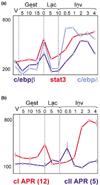
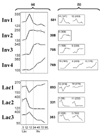
Comment in
-
Evolving views of involution.Breast Cancer Res. 2004;6(2):89-92. doi: 10.1186/bcr765. Epub 2004 Feb 10. Breast Cancer Res. 2004. PMID: 14979913 Free PMC article. Review.
Similar articles
-
Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3.Breast Cancer Res. 2004;6(2):R75-91. doi: 10.1186/bcr753. Epub 2003 Dec 18. Breast Cancer Res. 2004. PMID: 14979920 Free PMC article.
-
Immune cell regulators in mouse mammary development and involution.J Anim Sci. 2009 Apr;87(13 Suppl):35-42. doi: 10.2527/jas.2008-1333. Epub 2008 Oct 10. J Anim Sci. 2009. PMID: 18849387 Review.
-
Overexpression and forced activation of stat5 in mammary gland of transgenic mice promotes cellular proliferation, enhances differentiation, and delays postlactational apoptosis.Mol Cancer Res. 2002 Nov;1(1):32-47. Mol Cancer Res. 2002. PMID: 12496367
-
Deletion of Stat3 blocks mammary gland involution and extends functional competence of the secretory epithelium in the absence of lactogenic stimuli.Endocrinology. 2002 Sep;143(9):3641-50. doi: 10.1210/en.2002-220224. Endocrinology. 2002. PMID: 12193580
-
The role of Stat3 in apoptosis and mammary gland involution. Conditional deletion of Stat3.Adv Exp Med Biol. 2000;480:129-38. doi: 10.1007/0-306-46832-8_16. Adv Exp Med Biol. 2000. PMID: 10959419 Review.
Cited by
-
Insight into mammary gland development and tumor progression in an E2F5 conditional knockout mouse model.Oncogene. 2024 Sep 28. doi: 10.1038/s41388-024-03172-4. Online ahead of print. Oncogene. 2024. PMID: 39341991
-
Immune Cell Contribution to Mammary Gland Development.J Mammary Gland Biol Neoplasia. 2024 Aug 23;29(1):16. doi: 10.1007/s10911-024-09568-y. J Mammary Gland Biol Neoplasia. 2024. PMID: 39177859 Free PMC article. Review.
-
Altered liver metabolism post-wean abolishes efficacy of vitamin D for breast cancer prevention in a mouse model.bioRxiv [Preprint]. 2024 Jun 2:2024.05.28.596304. doi: 10.1101/2024.05.28.596304. bioRxiv. 2024. PMID: 38854129 Free PMC article. Preprint.
-
Methods for investigating STAT3 regulation of lysosomal function in mammary epithelial cells.J Mammary Gland Biol Neoplasia. 2024 May 18;29(1):11. doi: 10.1007/s10911-024-09563-3. J Mammary Gland Biol Neoplasia. 2024. PMID: 38761238 Free PMC article.
-
In the Murine and Bovine Maternal Mammary Gland Signal Transducer and Activator of Transcription 3 is Activated in Clusters of Epithelial Cells around the Day of Birth.J Mammary Gland Biol Neoplasia. 2024 May 9;29(1):10. doi: 10.1007/s10911-024-09561-5. J Mammary Gland Biol Neoplasia. 2024. PMID: 38722417 Free PMC article.
References
-
- Strange R, Li F, Saurer S, Burkhardt A, Friis RR. Apoptotic cell death and tissue remodelling during mouse mammary gland involution. Development. 1992;115:49–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

