Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities
- PMID: 14963115
- PMCID: PMC369231
- DOI: 10.1128/jvi.78.5.2187-2200.2004
Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities
Abstract
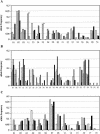
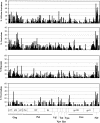
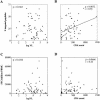
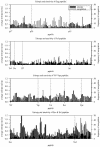
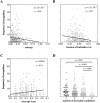
Although there is increasing evidence that virus-specific cytotoxic-T-lymphocyte (CTL) responses play an important role in the control of human immunodeficiency virus (HIV) replication in vivo, only scarce CTL data are available for the ethnic populations currently most affected by the epidemic. In this study, we examined the CD8(+)-T-cell responses in African-American, Caucasian, Hispanic, and Caribbean populations in which clade B virus dominates and analyzed the potential factors influencing immune recognition. Total HIV-specific CD8(+)-T-cell responses were determined by enzyme-linked immunospot assays in 150 HIV-infected individuals by using a clade B consensus sequence peptide set spanning all HIV proteins. A total of 88% of the 410 tested peptides were recognized, and Nef- and Gag-specific responses dominated the total response for each ethnicity in terms of both breadth and magnitude. Three dominantly targeted regions within these proteins that were recognized by >90% of individuals in each ethnicity were identified. Overall, the total breadth and magnitude of CD8(+)-T-cell responses correlated with individuals' CD4 counts but not with viral loads. The frequency of recognition for each peptide was highly correlated with the relative conservation of the peptide sequence, the presence of predicted immunoproteasomal cleavage sites within the C-terminal half of the peptide, and a reduced frequency of amino acids that impair binding of optimal epitopes to the restricting class I molecules. The present study thus identifies factors that contribute to the immunogenicity of these highly targeted and relatively conserved sequences in HIV that may represent promising vaccine candidates for ethnically heterogeneous populations.
Figures
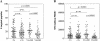
Similar articles
-
Differential narrow focusing of immunodominant human immunodeficiency virus gag-specific cytotoxic T-lymphocyte responses in infected African and caucasoid adults and children.J Virol. 2000 Jun;74(12):5679-90. doi: 10.1128/jvi.74.12.5679-5690.2000. J Virol. 2000. PMID: 10823876 Free PMC article.
-
Cytolytic T lymphocytes (CTLs) from HIV-1 subtype C-infected Indian patients recognize CTL epitopes from a conserved immunodominant region of HIV-1 Gag and Nef.J Infect Dis. 2005 Sep 1;192(5):749-59. doi: 10.1086/432547. Epub 2005 Jul 27. J Infect Dis. 2005. PMID: 16088824
-
HIV-1-specific cytotoxic T lymphocyte (CTL) responses against immunodominant optimal epitopes slow the progression of AIDS in China.Curr HIV Res. 2008 Jun;6(4):335-50. doi: 10.2174/157016208785132473. Curr HIV Res. 2008. PMID: 18691032
-
Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines.AIDS Res Hum Retroviruses. 2000 Sep 20;16(14):1433-43. doi: 10.1089/08892220050140982. AIDS Res Hum Retroviruses. 2000. PMID: 11018863 Review.
-
Characteristics of the intrahepatic cytotoxic T lymphocyte response in chronic hepatitis C virus infection.Springer Semin Immunopathol. 1997;19(1):69-83. doi: 10.1007/BF00945026. Springer Semin Immunopathol. 1997. PMID: 9266632 Review.
Cited by
-
Safety and immunogenicity of an HIV-1 gag DNA vaccine with or without IL-12 and/or IL-15 plasmid cytokine adjuvant in healthy, HIV-1 uninfected adults.PLoS One. 2012;7(1):e29231. doi: 10.1371/journal.pone.0029231. Epub 2012 Jan 5. PLoS One. 2012. PMID: 22242162 Free PMC article. Clinical Trial.
-
HLA alleles associated with slow progression to AIDS truly prefer to present HIV-1 p24.PLoS One. 2007 Sep 19;2(9):e920. doi: 10.1371/journal.pone.0000920. PLoS One. 2007. PMID: 17878955 Free PMC article.
-
Broad cross-clade T-cell responses to gag in individuals infected with human immunodeficiency virus type 1 non-B clades (A to G): importance of HLA anchor residue conservation.J Virol. 2005 Sep;79(17):11247-58. doi: 10.1128/JVI.79.17.11247-11258.2005. J Virol. 2005. PMID: 16103177 Free PMC article.
-
Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1.PLoS Pathog. 2007 Jul;3(7):e94. doi: 10.1371/journal.ppat.0030094. PLoS Pathog. 2007. PMID: 17616974 Free PMC article.
-
Fully differentiated HIV-1 specific CD8+ T effector cells are more frequently detectable in controlled than in progressive HIV-1 infection.PLoS One. 2007 Mar 28;2(3):e321. doi: 10.1371/journal.pone.0000321. PLoS One. 2007. PMID: 17389912 Free PMC article.
References
-
- Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. R. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. - PMC - PubMed
-
- Altfeld, M., M. M. Addo, R. Shankarappa, P. K. Lee, T. M. Allen, X. G. Yu, A. Rathod, J. D. Harlow, K. O'Sullivan, M. N. Johnston, P. J. R. Goulder, J. I. Mullins, E. S. Rosenberg, C. Brander, B. T. Korber, and B. D. Walker. 2003. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J. Virol. 77:7330-7340. - PMC - PubMed
-
- Altfeld, M., T. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. D. Harlow, P. J. R. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broadly directed pre-existing CD8+ T cell responses containing replication of the primary virus. Nature 420:434-439. - PubMed
-
- Altfeld, M., J. van Lunzen, N. Frahm, X. G. Yu, C. Schneider, R. L. Eldridge, M. E. Feeney, D. Meyer-Olson, H. J. Stellbrink, and B. D. Walker. 2002. Expansion of pre-existing, lymph node-localized CD8+ T cells during supervised treatment interruptions in chronic HIV-1 infection. J. Clin. Investig. 109:837-843. - PMC - PubMed
-
- Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

