Histone H3.3 is enriched in covalent modifications associated with active chromatin
- PMID: 14732680
- PMCID: PMC341768
- DOI: 10.1073/pnas.0308092100
Histone H3.3 is enriched in covalent modifications associated with active chromatin
Abstract
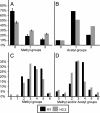
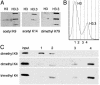
Chromatin states can be distinguished by differential covalent modifications of histones or by utilization of histone variants. Chromatin associated with transcriptionally active loci becomes enriched for histones with particular lysine modifications and accumulates the H3.3 histone variant, the substrate for replication-independent nucleosome assembly. However, studies of modifications at particular loci have not distinguished between histone variants, so the relationship among modifications, histone variants, and nucleosome assembly pathways is unclear. To address this uncertainty, we have quantified the relative abundance of H3 and H3.3 and their lysine modifications. Using a Drosophila cell line system in which H3.3 has been shown to specifically package active loci, we found that H3.3 accounts for approximately 25% of total histone 3 in bulk chromatin, enough to package essentially all actively transcribed genes. MS and antibody characterization of separated histone 3 fractions revealed that H3.3 is relatively enriched in modifications associated with transcriptional activity and deficient in dimethyl lysine-9, which is abundant in heterochromatin. To explain enrichment on alternative variants, we propose that histone modifications are tied to the alternative nucleosome assembly pathways that use primarily H3 at replication forks and H3.3 at actively transcribed genes in a replication-independent manner.
Figures
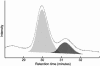
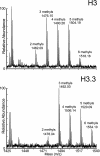
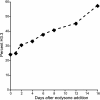
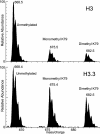
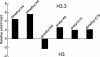
Comment in
-
Histone H3 variants and modifications on transcribed genes.Proc Natl Acad Sci U S A. 2004 Feb 10;101(6):1429-30. doi: 10.1073/pnas.0308506101. Epub 2004 Feb 2. Proc Natl Acad Sci U S A. 2004. PMID: 14757811 Free PMC article. No abstract available.
Similar articles
-
Plasticity of histone modifications across the invertebrate to vertebrate transition: histone H3 lysine 4 trimethylation in heterochromatin.Chromosome Res. 2005;13(1):57-72. doi: 10.1007/s10577-005-6845-6. Chromosome Res. 2005. PMID: 15791412
-
Histone H3 variants specify modes of chromatin assembly.Proc Natl Acad Sci U S A. 2002 Dec 10;99 Suppl 4(Suppl 4):16477-84. doi: 10.1073/pnas.172403699. Epub 2002 Aug 12. Proc Natl Acad Sci U S A. 2002. PMID: 12177448 Free PMC article. Review.
-
Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias.Genes Dev. 2005 Aug 1;19(15):1761-6. doi: 10.1101/gad.347705. Genes Dev. 2005. PMID: 16077006 Free PMC article.
-
Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly.Science. 2010 Apr 2;328(5974):94-8. doi: 10.1126/science.1178994. Science. 2010. PMID: 20360108
-
In and out: histone variant exchange in chromatin.Trends Biochem Sci. 2005 Dec;30(12):680-7. doi: 10.1016/j.tibs.2005.10.003. Epub 2005 Oct 28. Trends Biochem Sci. 2005. PMID: 16257529 Review.
Cited by
-
Proteomics in epigenetics: new perspectives for cancer research.Brief Funct Genomics. 2013 May;12(3):205-18. doi: 10.1093/bfgp/elt002. Epub 2013 Feb 11. Brief Funct Genomics. 2013. PMID: 23401080 Free PMC article. Review.
-
Incorporation of histone H3.1 suppresses the lineage potential of skeletal muscle.Nucleic Acids Res. 2015 Jan;43(2):775-86. doi: 10.1093/nar/gku1346. Epub 2014 Dec 24. Nucleic Acids Res. 2015. PMID: 25539924 Free PMC article.
-
Multiple Roles of dXNP and dADD1-Drosophila Orthologs of ATRX Chromatin Remodeler.Int J Mol Sci. 2023 Nov 18;24(22):16486. doi: 10.3390/ijms242216486. Int J Mol Sci. 2023. PMID: 38003676 Free PMC article. Review.
-
Lysine-36 of Drosophila histone H3.3 supports adult longevity.bioRxiv [Preprint]. 2023 Dec 13:2023.09.28.559962. doi: 10.1101/2023.09.28.559962. bioRxiv. 2023. Update in: G3 (Bethesda). 2024 Apr 3;14(4):jkae030. doi: 10.1093/g3journal/jkae030 PMID: 38196611 Free PMC article. Updated. Preprint.
-
Histone exchange, chromatin structure and the regulation of transcription.Nat Rev Mol Cell Biol. 2015 Mar;16(3):178-89. doi: 10.1038/nrm3941. Epub 2015 Feb 4. Nat Rev Mol Cell Biol. 2015. PMID: 25650798 Review.
References
-
- Workman, J. L. & Kingston, R. E. (1998) Annu. Rev. Biochem. 67, 545-579. - PubMed
-
- Weintraub, H. & Groudine, M. (1976) Science 193, 848-856. - PubMed
-
- Ahmad, K. & Henikoff, S. (2002) Mol. Cell 9, 1191-1200. - PubMed
-
- Smith, M. M. (2002) Mol. Cell 9, 1158-1160. - PubMed
-
- Turner, B. M. (2002) Cell 111, 285-291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

