L-Myc protein synthesis is initiated by internal ribosome entry
- PMID: 14730027
- PMCID: PMC1370540
- DOI: 10.1261/rna.5138804
L-Myc protein synthesis is initiated by internal ribosome entry
Abstract
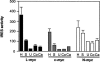
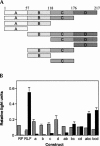
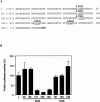
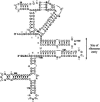
An internal ribosome entry segment (IRES) has been identified in the 5' untranslated region (5' UTR) of two members of the myc family of proto-oncogenes, c-myc and N-myc. Hence, the synthesis of c-Myc and N-Myc polypeptides can involve the alternative mechanism of internal initiation. Here, we show that the 5' UTR of L-myc, another myc family member, also contains an IRES. Previous studies have shown that the translation of mRNAs containing the c-myc and N-myc IRESs can involve both cap-dependent initiation and internal initiation. In contrast, the data presented here suggest that internal initiation can account for all of the translation initiation that occurs on an mRNA with the L-myc IRES in its 5' UTR. Like many other cellular IRESs, the L-myc IRES appears to be modular in nature and the entire 5' UTR is required for maximum IRES efficiency. The ribosome entry window within the L-myc IRES is located some distance upstream of the initiation codon, and thus, this IRES uses a "land and scan" mechanism to initiate translation. Finally, we have derived a secondary structural model for the IRES. The model confirms that the L-myc IRES is highly structured and predicts that a pseudoknot may form near the 5' end of the mRNA.
Figures

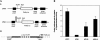
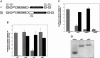

Similar articles
-
Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment.Nucleic Acids Res. 2000 Feb 1;28(3):687-94. doi: 10.1093/nar/28.3.687. Nucleic Acids Res. 2000. PMID: 10637319 Free PMC article.
-
C-Myc 5' untranslated region contains an internal ribosome entry segment.Oncogene. 1998 Jan 22;16(3):423-8. doi: 10.1038/sj.onc.1201763. Oncogene. 1998. PMID: 9467968
-
c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis.Mol Cell Biol. 2000 Feb;20(4):1162-9. doi: 10.1128/MCB.20.4.1162-1169.2000. Mol Cell Biol. 2000. PMID: 10648601 Free PMC article.
-
An atypical IRES within the 5' UTR of a dicistrovirus genome.Virus Res. 2009 Feb;139(2):157-65. doi: 10.1016/j.virusres.2008.07.017. Epub 2008 Sep 11. Virus Res. 2009. PMID: 18755228 Review.
-
Reversal of G-Quadruplexes' Role in Translation Control When Present in the Context of an IRES.Biomolecules. 2022 Feb 16;12(2):314. doi: 10.3390/biom12020314. Biomolecules. 2022. PMID: 35204814 Free PMC article. Review.
Cited by
-
Searching for IRES.RNA. 2006 Oct;12(10):1755-85. doi: 10.1261/rna.157806. Epub 2006 Sep 6. RNA. 2006. PMID: 16957278 Free PMC article. Review.
-
An miRNA-mediated therapy for SCA6 blocks IRES-driven translation of the CACNA1A second cistron.Sci Transl Med. 2016 Jul 13;8(347):347ra94. doi: 10.1126/scitranslmed.aaf5660. Sci Transl Med. 2016. PMID: 27412786 Free PMC article.
-
The dicistronic RNA from the mouse LINE-1 retrotransposon contains an internal ribosome entry site upstream of each ORF: implications for retrotransposition.Nucleic Acids Res. 2006 Feb 6;34(3):853-64. doi: 10.1093/nar/gkj490. Print 2006. Nucleic Acids Res. 2006. PMID: 16464823 Free PMC article.
-
Translational Regulations in Response to Endoplasmic Reticulum Stress in Cancers.Cells. 2020 Feb 26;9(3):540. doi: 10.3390/cells9030540. Cells. 2020. PMID: 32111004 Free PMC article. Review.
-
The genetic code as expressed through relationships between mRNA structure and protein function.FEBS Lett. 2013 Apr 17;587(8):1180-1188. doi: 10.1016/j.febslet.2013.03.002. Epub 2013 Mar 13. FEBS Lett. 2013. PMID: 23499436 Free PMC article. Review.
References
-
- Belsham, G.J. and Jackson, R.J. 2000. Translation initiation on Picornavirus RNA. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 869–900. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Birrer, M.J., Dosaka, H., Segal, S., Nau, M., Degreve, J., Kaye, F., Sausville, E., and Minna, J. 1988a. Multiple L-myc messenger-RNA forms have transforming activity for primary rat embryo fibroblasts and an established rat fibroblast cell-line. Clinical Res. 36: A589–A589.
-
- Blackwood, E.M., Luscher, B., and Eisenman, R.N. 1992. Myc and Max associate in vivo. Genes & Dev. 6: 71–80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
