IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding
- PMID: 14657035
- PMCID: PMC291812
- DOI: 10.1093/emboj/cdg617
IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding
Abstract
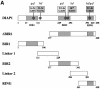
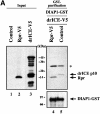
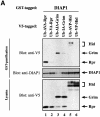
The Drosophila inhibitor of apoptosis protein DIAP1 ensures cell viability by directly inhibiting caspases. In cells destined to die this IAP-mediated inhibition of caspases is overcome by IAP-antagonists. Genetic evidence indicates that IAP-antagonists are non-equivalent and function synergistically to promote apoptosis. Here we provide biochemical evidence for the non-equivalent mode of action of Reaper, Grim, Hid and Jafrac2. We find that these IAP-antagonists display differential and selective binding to specific DIAP1 BIR domains. Consistently, we show that each DIAP1 BIR region associates with distinct caspases. The differential DIAP1 BIR interaction seen both between initiator and effector caspases and within IAP-antagonist family members suggests that different IAP-antagonists inhibit distinct caspases from interacting with DIAP1. Surprisingly, we also find that the caspase-binding residues of XIAP predicted to be strictly conserved in caspase-binding IAPs, are absent in DIAP1. In contrast to XIAP, residues C-terminal to the DIAP1 BIR1 domain are indispensable for caspase association. Our studies on DIAP1 and caspases expose significant differences between DIAP1 and XIAP suggesting that DIAP1 and XIAP inhibit caspases in different ways.
Figures



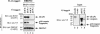
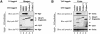
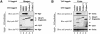




Similar articles
-
Jafrac2 is an IAP antagonist that promotes cell death by liberating Dronc from DIAP1.EMBO J. 2002 Oct 1;21(19):5118-29. doi: 10.1093/emboj/cdf530. EMBO J. 2002. PMID: 12356728 Free PMC article.
-
The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers.J Biol Chem. 2007 Jan 19;282(3):2056-68. doi: 10.1074/jbc.M608051200. Epub 2006 Oct 26. J Biol Chem. 2007. PMID: 17068333
-
Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.Nat Struct Mol Biol. 2004 May;11(5):420-8. doi: 10.1038/nsmb764. Epub 2004 Apr 25. Nat Struct Mol Biol. 2004. PMID: 15107838
-
Regulation of Cell Death by IAPs and Their Antagonists.Curr Top Dev Biol. 2015;114:185-208. doi: 10.1016/bs.ctdb.2015.07.026. Epub 2015 Sep 11. Curr Top Dev Biol. 2015. PMID: 26431568 Free PMC article. Review.
-
Regulation of apoptosis in Drosophila.Cell Death Differ. 2008 Jul;15(7):1132-8. doi: 10.1038/cdd.2008.50. Epub 2008 Apr 25. Cell Death Differ. 2008. PMID: 18437164 Review.
Cited by
-
The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila.Development. 2005 May;132(9):2125-34. doi: 10.1242/dev.01790. Epub 2005 Mar 30. Development. 2005. PMID: 15800001 Free PMC article.
-
Drosophila caspases as guardians of host-microbe interactions.Cell Death Differ. 2023 Feb;30(2):227-236. doi: 10.1038/s41418-022-01038-4. Epub 2022 Jul 9. Cell Death Differ. 2023. PMID: 35810247 Free PMC article. Review.
-
Drosophila USP5 controls the activation of apoptosis and the Jun N-terminal kinase pathway during eye development.PLoS One. 2014 Mar 18;9(3):e92250. doi: 10.1371/journal.pone.0092250. eCollection 2014. PLoS One. 2014. PMID: 24643212 Free PMC article.
-
Genetic control of programmed cell death (apoptosis) in Drosophila.Fly (Austin). 2009 Jan-Mar;3(1):78-90. doi: 10.4161/fly.3.1.7800. Epub 2009 Jan 8. Fly (Austin). 2009. PMID: 19182545 Free PMC article. Review.
-
A lepidopteran orthologue of reaper reveals functional conservation and evolution of IAP antagonists.Insect Mol Biol. 2009 Jun;18(3):341-51. doi: 10.1111/j.1365-2583.2009.00878.x. Insect Mol Biol. 2009. PMID: 19523066 Free PMC article.
References
-
- Chai J., Du,C., Wu,J.W., Kyin,S., Wang,X. and Shi,Y. (2000) Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature, 406, 855–862. - PubMed
-
- Chen P., Nordstrom,W., Gish,B. and Abrams,J.M. (1996) grim, a novel cell death gene in Drosophila. Genes Dev., 10, 1773–1782. - PubMed
-
- Christich A., Kauppila,S., Chen,P., Sogame,N., Ho,S.I. and Abrams,J.M. (2002) The damage-responsive Drosophila gene sickle encodes a novel IAP binding protein similar to but distinct from reaper, grim and hid. Curr. Biol., 12, 137–140. - PubMed
-
- Ditzel M., Wilson,R., Tenev,T., Zachariou,A., Paul,A., Deas,E. and Meier,P. (2003) Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell Biol., 5, 467–473. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

