Folate receptor alpha and caveolae are not required for Ebola virus glycoprotein-mediated viral infection
- PMID: 14645601
- PMCID: PMC296046
- DOI: 10.1128/jvi.77.24.13433-13438.2003
Folate receptor alpha and caveolae are not required for Ebola virus glycoprotein-mediated viral infection
Abstract
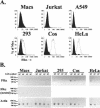
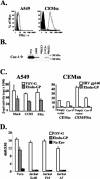
Folate receptor alpha (FRalpha) has been described as a factor involved in mediating Ebola virus entry into cells (6). Furthermore, it was suggested that interaction with FRalpha results in internalization and subsequent viral ingress into the cytoplasm via caveolae (9). Descriptions of cellular receptors for Ebola virus and its entry mechanisms are of fundamental importance, particularly with the advent of vectors bearing Ebola virus glycoprotein (GP) being utilized for gene transfer into cell types such as airway epithelial cells. Thus, the ability of FRalpha to mediate efficient entry of viral pseudotypes carrying GP was investigated. We identified cell lines and primary cell types such as macrophages that were readily infected by GP pseudotypes despite lacking detectable surface FRalpha, indicating that this receptor is not essential for Ebola virus infection. Furthermore, we find that T-cell lines stably expressing FRalpha are not infectible, suggesting that FRalpha is also not sufficient to mediate entry. T-cell lines lack caveolae, the predominant route of FRalpha-mediated folate metabolism. However, the coexpression of FRalpha with caveolin-1, the major structural protein of caveolae, was not able to rescue infectivity in a T-cell line. In addition, other cell types lacking caveolae are fully infectible by GP pseudotypes. Finally, a panel of ligands to and soluble analogues of FRalpha were unable to inhibit infection on a range of cell lines, questioning the role of FRalpha as an important factor for Ebola virus entry.
Figures
Similar articles
-
Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines.J Virol. 1998 Apr;72(4):3155-60. doi: 10.1128/JVI.72.4.3155-3160.1998. J Virol. 1998. PMID: 9525641 Free PMC article.
-
Characterization of Influenza Virus Pseudotyped with Ebolavirus Glycoprotein.J Virol. 2018 Jan 30;92(4):e00941-17. doi: 10.1128/JVI.00941-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29212933 Free PMC article.
-
A comparison of caveolae and caveolin-1 to folate receptor alpha in retina and retinal pigment epithelium.Histochem J. 2001 Mar;33(3):149-58. doi: 10.1023/a:1017991925821. Histochem J. 2001. PMID: 11508338 Free PMC article.
-
The role of folate receptor alpha in cancer development, progression and treatment: cause, consequence or innocent bystander?Int J Cancer. 2006 Jul 15;119(2):243-50. doi: 10.1002/ijc.21712. Int J Cancer. 2006. PMID: 16453285 Review.
-
[Ebola virus host cell entry].Uirusu. 2015;65(1):71-82. doi: 10.2222/jsv.65.71. Uirusu. 2015. PMID: 26923960 Review. Japanese.
Cited by
-
Comprehensive analysis of ebola virus GP1 in viral entry.J Virol. 2005 Apr;79(8):4793-805. doi: 10.1128/JVI.79.8.4793-4805.2005. J Virol. 2005. PMID: 15795265 Free PMC article.
-
Role of Clathrin Assembly Protein-2 Beta Subunit during White Spot Syndrome Virus Infection in Black Tiger Shrimp Penaeus monodon.Sci Rep. 2019 Sep 17;9(1):13489. doi: 10.1038/s41598-019-49852-0. Sci Rep. 2019. PMID: 31530841 Free PMC article.
-
The Ebola virus glycoprotein mediates entry via a non-classical dynamin-dependent macropinocytic pathway.Virology. 2011 Oct 25;419(2):72-83. doi: 10.1016/j.virol.2011.08.009. Epub 2011 Sep 9. Virology. 2011. PMID: 21907381 Free PMC article.
-
An update on the use of antibodies against the filoviruses.Immunotherapy. 2013 Nov;5(11):1221-33. doi: 10.2217/imt.13.124. Immunotherapy. 2013. PMID: 24188676 Free PMC article. Review.
-
Sphingosine Kinases Promote Ebola Virus Infection and Can Be Targeted to Inhibit Filoviruses, Coronaviruses, and Arenaviruses Using Late Endocytic Trafficking to Enter Cells.ACS Infect Dis. 2023 May 12;9(5):1064-1077. doi: 10.1021/acsinfecdis.2c00416. Epub 2023 Apr 13. ACS Infect Dis. 2023. PMID: 37053583 Free PMC article.
References
-
- Baskerville, A., S. P. Fisher-Hoch, G. H. Neild, and A. B. Dowsett. 1985. Ultrastructural pathology of experimental Ebola haemorrhagic fever virus infection. J. Pathol. 147:199-209. - PubMed
-
- Brzezinska, A., P. Winska, and M. Balinska. 2000. Cellular aspects of folate and antifolate membrane transport. Acta Biochim. Pol. 47:735-749. - PubMed
-
- Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151-1154. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

