Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome
- PMID: 14581452
- PMCID: PMC2173515
- DOI: 10.1083/jcb.200305007
Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome
Abstract
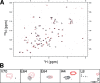
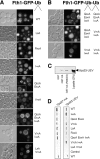
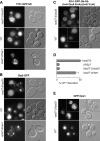
Ubiquitin (Ub) attachment to cell surface proteins causes their lysosomal degradation by incorporating them into lumenal membranes of multivesicular bodies (MVBs). Two yeast endosomal protein complexes have been proposed as Ub-sorting "receptors," the Vps27-Hse1 complex and the ESCRT-I complex. We used NMR spectroscopy and mutagenesis studies to map the Ub-binding surface for Vps27 and Vps23. Mutations in Ub that ablate only Vps27 binding or Vps23 binding blocked the ability of Ub to serve as an MVB sorting signal, supporting the idea that both the Vps27-Hse1 and ESCRT-I complexes interact with ubiquitinated cargo. Vps27 also bound Vps23 directly via two PSDP motifs present within the Vps27 COOH terminus. Loss of Vps27-Vps23 association led to less efficient sorting into the endosomal lumen. However, sorting of vacuolar proteases or the overall biogenesis of the MVB were not grossly affected. In contrast, disrupting interaction between Vps27 and Hse1 caused severe defects in carboxy peptidase Y sorting and MVB formation. These results indicate that both Ub-sorting complexes are coupled for efficient recognition of ubiquitinated cargo.
Figures



Similar articles
-
Vps27 recruits ESCRT machinery to endosomes during MVB sorting.J Cell Biol. 2003 Aug 4;162(3):413-23. doi: 10.1083/jcb.200302136. J Cell Biol. 2003. PMID: 12900393 Free PMC article.
-
Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins.J Biol Chem. 2004 Jul 2;279(27):28689-96. doi: 10.1074/jbc.M400023200. Epub 2004 Mar 24. J Biol Chem. 2004. PMID: 15044434
-
Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I.Cell. 2001 Jul 27;106(2):145-55. doi: 10.1016/s0092-8674(01)00434-2. Cell. 2001. PMID: 11511343
-
Protein sorting into multivesicular endosomes.Curr Opin Cell Biol. 2003 Aug;15(4):446-55. doi: 10.1016/s0955-0674(03)00080-2. Curr Opin Cell Biol. 2003. PMID: 12892785 Review.
-
Ubiquitin and endocytic protein sorting.Essays Biochem. 2005;41:81-98. doi: 10.1042/EB0410081. Essays Biochem. 2005. PMID: 16250899 Review.
Cited by
-
Solution structure of the ESCRT-I and -II supercomplex: implications for membrane budding and scission.Structure. 2012 May 9;20(5):874-86. doi: 10.1016/j.str.2012.03.008. Structure. 2012. PMID: 22579254 Free PMC article.
-
Multiomics of GCN4-Dependent Replicative Lifespan Extension Models Reveals Gcn4 as a Regulator of Protein Turnover in Yeast.Int J Mol Sci. 2023 Nov 10;24(22):16163. doi: 10.3390/ijms242216163. Int J Mol Sci. 2023. PMID: 38003352 Free PMC article.
-
The ubiquitin ligase UBE4B regulates amyloid precursor protein ubiquitination, endosomal trafficking, and amyloid β42 generation and secretion.Mol Cell Neurosci. 2020 Oct;108:103542. doi: 10.1016/j.mcn.2020.103542. Epub 2020 Aug 22. Mol Cell Neurosci. 2020. PMID: 32841720 Free PMC article.
-
Pkh1/2-dependent phosphorylation of Vps27 regulates ESCRT-I recruitment to endosomes.Mol Biol Cell. 2012 Oct;23(20):4054-64. doi: 10.1091/mbc.E12-01-0001. Epub 2012 Aug 23. Mol Biol Cell. 2012. PMID: 22918958 Free PMC article.
-
Role of Ceramides and Lysosomes in Extracellular Vesicle Biogenesis, Cargo Sorting and Release.Int J Mol Sci. 2022 Dec 5;23(23):15317. doi: 10.3390/ijms232315317. Int J Mol Sci. 2022. PMID: 36499644 Free PMC article. Review.
References
-
- Babst, M., G. Odorizzi, E.J. Estepa, and S.D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 1:248–258. - PubMed
-
- Babst, M., D.J. Katzmann, W.B. Snyder, B. Wendland, and S.D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 3:283–289. - PubMed
-
- Bache, K.G., C. Raiborg, A. Mehlum, and H. Stenmark. 2003. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278:12513–12521. - PubMed
-
- Bilodeau, P.S., J.L. Urbanowski, S.C. Winistorfer, and R.C. Piper. 2002. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4:534–539. - PubMed
-
- Carter, C.A. 2002. Tsg101: HIV-1's ticket to ride. Trends Microbiol. 10:203–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

